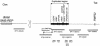The evolutionary chromosome translocation 4;19 in Gorilla gorilla is associated with microduplication of the chromosome fragment syntenic to sequences surrounding the human proximal CMT1A-REP - PubMed (original) (raw)
The evolutionary chromosome translocation 4;19 in Gorilla gorilla is associated with microduplication of the chromosome fragment syntenic to sequences surrounding the human proximal CMT1A-REP
P Stankiewicz et al. Genome Res. 2001 Jul.
Abstract
Many genomic disorders occur as a result of chromosome rearrangements involving low-copy repeats (LCRs). To better understand the molecular basis of chromosome rearrangements, including translocations, we have investigated the mechanism of evolutionary rearrangements. In contrast to several intrachromosomal rearrangements, only two evolutionary translocations have been identified by cytogenetic analyses of humans and greater apes. Human chromosome 2 arose as a result of a telomeric fusion between acrocentric chromosomes, whereas chromosomes 4 and 19 in Gorilla gorilla are the products of a reciprocal translocation between ancestral chromosomes, syntenic to human chromosomes 5 and 17, respectively. Fluorescence in situ hybridization (FISH) was used to characterize the breakpoints of the latter translocation at the molecular level. We identified three BAC clones that span translocation breakpoints. One breakpoint occurred in the region syntenic to human chromosome 5q13.3, between the HMG-CoA reductase gene (HMGCR) and RAS p21 protein activator 1 gene (RASA1). The second breakpoint was in a region syntenic to human chromosome 17p12 containing the 24 kb region-specific low-copy repeat-proximal CMT1A-REP. Moreover, we found that the t(4;19) is associated with a submicroscopic chromosome duplication involving a 19p chromosome fragment homologous to the human chromosome region surrounding the proximal CMT1A-REP. These observations further indicate that higher order genomic architecture involving low-copy repeats resulting from genomic duplication plays a significant role in karyotypic evolution.
Figures
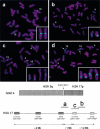
Figure 1
Gorilla metaphase chromosomes after hybridization with human BAC clones. Below is shown gorilla chromosome 4 (GGO 4), syntenic to human chromosomes 5 and 17. The vertical arrow indicates the ∼250 kb genomic duplication, originating from the GGO 19 segment, syntenic to human chromosome, (HSA) 17p12. Smith-Magenis syndrome low-copy repeats, SMS-REPs, and Charcot-Marie-Tooth disease type 1A low copy repeats, CMT1A-REPs, in the human chromosome 17p11.2p12 region are shown (telomere to right of figure) with the relative physical position of three BAC clones used in the FISH analysis that is shown above. (a) CTD-3157E16 (HSA 17p12), located proximal to the translocation breakpoint. (b) RP11–726O12 (HSA 17p12), located distal to the translocation breakpoint. (c) RP11-385D13 (HSA 17p12, proximal CMT1A-REP), spans the translocation breakpoint and includes the genomic duplication in gorilla. (d) CTC-428H11 (HSA 5q13.3) spans the breakpoint. Arrows within the FISH picture indicate an additional hybridization signal, which is also located on the long arm of GGO 19. This apparent duplication is located distal to the evolutionary breakpoint identified in this study. Small panels in the lower right corners demonstrate only the gorilla chromosomes 4 and 19 with positive probe hybridization signals.
Figure 2
Schematic representation of the gorilla chromosome 19 duplicated region. Human BAC, PAC, and cosmid clones are shown below. Note that the proximal CMT1A-REP is not present in the gorilla genome. Its position in human is indicated with an arrow.
Similar articles
- Molecular evolution of the CMT1A-REP region: a human- and chimpanzee-specific repeat.
Keller MP, Seifried BA, Chance PF. Keller MP, et al. Mol Biol Evol. 1999 Aug;16(8):1019-26. doi: 10.1093/oxfordjournals.molbev.a026191. Mol Biol Evol. 1999. PMID: 10474898 - Evidence for involvement of TRE-2 (USP6) oncogene, low-copy repeat and acrocentric heterochromatin in two families with chromosomal translocations.
Ou Z, Jarmuz M, Sparagana SP, Michaud J, Décarie JC, Yatsenko SA, Nowakowska B, Furman P, Shaw CA, Shaffer LG, Lupski JR, Chinault AC, Cheung SW, Stankiewicz P. Ou Z, et al. Hum Genet. 2006 Sep;120(2):227-37. doi: 10.1007/s00439-006-0200-7. Epub 2006 Jun 22. Hum Genet. 2006. PMID: 16791615 - The 1.4-Mb CMT1A duplication/HNPP deletion genomic region reveals unique genome architectural features and provides insights into the recent evolution of new genes.
Inoue K, Dewar K, Katsanis N, Reiter LT, Lander ES, Devon KL, Wyman DW, Lupski JR, Birren B. Inoue K, et al. Genome Res. 2001 Jun;11(6):1018-33. doi: 10.1101/gr.180401. Genome Res. 2001. PMID: 11381029 Free PMC article.
Cited by
- Detection and analysis of ancient segmental duplications in mammalian genomes.
Pu L, Lin Y, Pevzner PA. Pu L, et al. Genome Res. 2018 Jun;28(6):901-909. doi: 10.1101/gr.228718.117. Epub 2018 May 7. Genome Res. 2018. PMID: 29735604 Free PMC article. - Genomic disorders ten years on.
Lupski JR. Lupski JR. Genome Med. 2009 Apr 24;1(4):42. doi: 10.1186/gm42. Genome Med. 2009. PMID: 19439022 Free PMC article. - Large-scale variation among human and great ape genomes determined by array comparative genomic hybridization.
Locke DP, Segraves R, Carbone L, Archidiacono N, Albertson DG, Pinkel D, Eichler EE. Locke DP, et al. Genome Res. 2003 Mar;13(3):347-57. doi: 10.1101/gr.1003303. Genome Res. 2003. PMID: 12618365 Free PMC article. - A genome-wide survey of structural variation between human and chimpanzee.
Newman TL, Tuzun E, Morrison VA, Hayden KE, Ventura M, McGrath SD, Rocchi M, Eichler EE. Newman TL, et al. Genome Res. 2005 Oct;15(10):1344-56. doi: 10.1101/gr.4338005. Epub 2005 Sep 16. Genome Res. 2005. PMID: 16169929 Free PMC article.
References
- Arnold N, Stanyon R, Jauch A, O'Brien P, Wienberg J. Identification of complex chromosome rearrangements in the gibbon by fluorescent in situ hybridization (FISH) of a human chromosome 2q specific microlibrary, yeast artificial chromosomes, and reciprocal chromosome painting. Cytogenet Cell Genet. 1996;74:80–85. - PubMed
- Boerkoel CF, Inoue K, Reiter LT, Warner LE, Lupski JR. Molecular mechanisms for CMT1A duplication and HNPP deletion. Ann N Y Acad Sci. 1999;883:22–35. - PubMed
- Chen K-S, Manian P, Koeuth T, Potocki L, Zhao Q, Chinault AC, Lee CC, Lupski JR. Homologous recombination of a flanking repeat gene cluster is a mechanism for a common contiguous gene deletion syndrome. Nat Genet. 1997;17:154–163. - PubMed
- Dutrillaux B. Chromosomal evolution in primates: Tentative phylogeny from Microcebus murinus (Prosimian) to man. Hum Genet. 1979;48:251–314. - PubMed
- Dutrillaux B, Rethore MO, Prieur M, Lejeune J. Analysis of the structure of chromatids of Gorilla gorilla. Comparison with Homo sapiens and Pan troglodytes. Humangenetik. 1973;20:343–354. - PubMed
Publication types
MeSH terms
Grants and funding
- P01 HD039420/HD/NICHD NIH HHS/United States
- R01 NS027042/NS/NINDS NIH HHS/United States
- P01 HD39420/HD/NICHD NIH HHS/United States
- R01 NS27042/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous