Location and properties of metal-binding sites on the human prion protein - PubMed (original) (raw)
Location and properties of metal-binding sites on the human prion protein
G S Jackson et al. Proc Natl Acad Sci U S A. 2001.
Abstract
Although a functional role in copper binding has been suggested for the prion protein, evidence for binding at affinities characteristic of authentic metal-binding proteins has been lacking. By presentation of copper(II) ions in the presence of the weak chelator glycine, we have now characterized two high-affinity binding sites for divalent transition metals within the human prion protein. One is in the N-terminal octapeptide-repeat segment and has a K(d) for copper(II) of 10(-14) M, with other metals (Ni(2+), Zn(2+), and Mn(2+)) binding three or more orders of magnitude more weakly. However, NMR and fluorescence data reveal a previously unreported second site around histidines 96 and 111, a region of the molecule known to be crucial for prion propagation. The K(d) for copper(II) at this site is 4 x 10(-14) M, whereas nickel(II), zinc(II), and manganese(II) bind 6, 7, and 10 orders of magnitude more weakly, respectively, regardless of whether the protein is in its oxidized alpha-helical (alpha-PrP) or reduced beta-sheet (beta-PrP) conformation. A role for prion protein (PrP) in copper metabolism or transport seems likely and disturbance of this function may be involved in prion-related neurotoxicity.
Figures
Figure 1
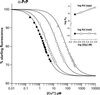
Metal binding to recombinant human α-PrP91–231. The main figure shows the data for the binding of copper(II) to human PrP91–231 in the presence of increasing concentrations of glycine. The quenching of intrinsic fluorescence from tryptophan-99 is shown as a percentage of the starting fluorescence and is plotted versus the concentration of copper(II) ions. The data in the presence of 100 μM glycine are shown by solid circles, 200 μM by open squares; 500 μM by open circles, and 1 mM by open triangles. The lines superimposed on the data represent an optimal fit of the data to the equation described above. (Inset) A plot of the apparent dissociation constants versus glycine concentration, which clearly vary. Overlaid are the real dissociation constants derived as described above, which are constant with respect to glycine concentration.
Figure 2
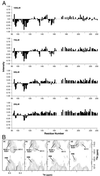
NMR signal intensity versus residue number. (A) HSQC peak intensities shown sequentially at 250 μM, 500 μM, 750 μM, and 1000 μM CuSO4 relative to 0 μM CuSO4; showing the effects of addition of Cu2+ to the human PrP91–231. Residues 94–98, 108–114, 135–136, and 153–159 exhibit significant differential alteration of resonances, indicating copper binding. (B) 1H-15N HSQC spectra with 0 μM (a), 375 μM (b), 625 μM (c), and 1000 μM CuSO4 (d), showing the region surrounding peaks arising from H96 and H111. As Cu2+ is titrated into the NMR sample, a general broadening of NMR resonances is observed, due to the presence of Cu2+ in solution. In addition, the peaks arising, for example, from the backbone amide resonances of H96 and H111 exhibit significant further broadening because of their proximity to the binding site of the paramagnetic ion. The peaks arising from the backbone amides of residues N197 and E221 are shown for comparison.
Figure 3
Binding of copper to the octapeptide repeat region PrP52–98. The data for the binding of copper(II) to PrP52–98 in both the presence of 500 μM glycine (open squares) and the absence of glycine (open circles) are shown overlaid. The horizontal dotted line is drawn at a point representing stoichiometry of copper(II) to PrP52–98 (5 μM) to illustrate the loss of the weak-binding site in the presence of glycine. The lines superimposed on the data represent an optimal fit of the data to the equation described above.
Figure 4
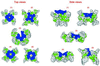
Five energetically identical space-filled models of the Cu(II)-binding octapeptide repeat region. Models were generated by calculating the energetic stabilities of all possible combinations of exactly repeating structure in the octapeptide repeat. To enable exhaustive structural evaluation within the repeating unit, a simplified energetic and structural model was applied. Energetic stabilities were calculated by using a united atom model and physicochemical potentials describing hydrophobic, hydrophilic, and backbone hydrogen bonding energies. Conformational space was reduced, by restricting residues to adopt one of six alternative backbone φ–ψ torsion angles. The system is described in detail elsewhere (21). The models presented represent the five most energetically stable conformations of the repeat that also satisfy the proposed coordination of the copper ion (yellow) by the histidine side chains (blue). The remaining side chains are colored gray and backbone atoms are represented by green.
Similar articles
- Interaction of the human prion PrP(106-126) sequence with copper(II), manganese(II), and zinc(II): NMR and EPR studies.
Gaggelli E, Bernardi F, Molteni E, Pogni R, Valensin D, Valensin G, Remelli M, Luczkowski M, Kozlowski H. Gaggelli E, et al. J Am Chem Soc. 2005 Jan 26;127(3):996-1006. doi: 10.1021/ja045958z. J Am Chem Soc. 2005. PMID: 15656638 - Mixed metal copper(II)-nickel(II) and copper(II)-zinc(II) complexes of multihistidine peptide fragments of human prion protein.
Jószai V, Turi I, Kállay C, Pappalardo G, Di Natale G, Rizzarelli E, Sóvágó I. Jószai V, et al. J Inorg Biochem. 2012 Jul;112:17-24. doi: 10.1016/j.jinorgbio.2012.02.014. Epub 2012 Feb 28. J Inorg Biochem. 2012. PMID: 22542592 - Real-time kinetics of discontinuous and highly conformational metal-ion binding sites of prion protein.
Treiber C, Thompsett AR, Pipkorn R, Brown DR, Multhaup G. Treiber C, et al. J Biol Inorg Chem. 2007 Jun;12(5):711-20. doi: 10.1007/s00775-007-0220-3. Epub 2007 Mar 8. J Biol Inorg Chem. 2007. PMID: 17345106 - Prion proteins leading to neurodegeneration.
La Mendola D, Pietropaolo A, Pappalardo G, Zannoni C, Rizzarelli E. La Mendola D, et al. Curr Alzheimer Res. 2008 Dec;5(6):579-90. doi: 10.2174/156720508786898415. Curr Alzheimer Res. 2008. PMID: 19075585 Review. - A Yin-Yang role for metals in prion disease.
Wong BS, Brown DR, Sy MS. Wong BS, et al. Panminerva Med. 2001 Dec;43(4):283-7. Panminerva Med. 2001. PMID: 11677424 Review.
Cited by
- Aβ neurotoxicity depends on interactions between copper ions, prion protein, and N-methyl-D-aspartate receptors.
You H, Tsutsui S, Hameed S, Kannanayakal TJ, Chen L, Xia P, Engbers JD, Lipton SA, Stys PK, Zamponi GW. You H, et al. Proc Natl Acad Sci U S A. 2012 Jan 31;109(5):1737-42. doi: 10.1073/pnas.1110789109. Epub 2012 Jan 17. Proc Natl Acad Sci U S A. 2012. PMID: 22307640 Free PMC article. - Prion diseases.
McKintosh E, Tabrizi SJ, Collinge J. McKintosh E, et al. J Neurovirol. 2003 Apr;9(2):183-93. doi: 10.1080/13550280390194082. J Neurovirol. 2003. PMID: 12707849 Review. - PrP (58-93) peptide from unstructured N-terminal domain of human prion protein forms amyloid-like fibrillar structures in the presence of Zn2+ ions.
Gielnik M, Pietralik Z, Zhukov I, Szymańska A, Kwiatek WM, Kozak M. Gielnik M, et al. RSC Adv. 2019 Jul 17;9(39):22211-22219. doi: 10.1039/c9ra01510h. eCollection 2019 Jul 17. RSC Adv. 2019. PMID: 35519468 Free PMC article. - Mouse brain synaptosomes accumulate copper-67 efficiently by two distinct processes independent of cellular prion protein.
Giese A, Buchholz M, Herms J, Kretzschmar HA. Giese A, et al. J Mol Neurosci. 2005;27(3):347-54. doi: 10.1385/JMN:27:3:347. J Mol Neurosci. 2005. PMID: 16280605 - Titanium dioxide and carbon black nanoparticles disrupt neuronal homeostasis via excessive activation of cellular prion protein signaling.
Ribeiro LW, Pietri M, Ardila-Osorio H, Baudry A, Boudet-Devaud F, Bizingre C, Arellano-Anaya ZE, Haeberlé AM, Gadot N, Boland S, Devineau S, Bailly Y, Kellermann O, Bencsik A, Schneider B. Ribeiro LW, et al. Part Fibre Toxicol. 2022 Jul 15;19(1):48. doi: 10.1186/s12989-022-00490-x. Part Fibre Toxicol. 2022. PMID: 35840975 Free PMC article.
References
- Jackson G S, Clarke A R. Curr Opin Struct Biol. 2000;10:69–74. - PubMed
- Stahl N, Baldwin M A, Teplow D B, Hood L, Gibson B W, Burlingame A L, Prusiner S B. Biochemistry. 1993;32:1991–2002. - PubMed
- Wadsworth J D F, Hill A F, Joiner S, Jackson G S, Clarke A R, Collinge J. Nat Cell Biol. 1999;1:55–59. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials