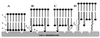TAT peptide on the surface of liposomes affords their efficient intracellular delivery even at low temperature and in the presence of metabolic inhibitors - PubMed (original) (raw)
TAT peptide on the surface of liposomes affords their efficient intracellular delivery even at low temperature and in the presence of metabolic inhibitors
V P Torchilin et al. Proc Natl Acad Sci U S A. 2001.
Abstract
To achieve an efficient intracellular drug and DNA delivery, attempts were made to target microparticulate drug carriers into cytoplasm bypassing the endocytotic pathway. TAT peptides derived from the HIV-1 TAT protein facilitate intracellular delivery of proteins and small colloidal particles. We demonstrated that relatively large drug carriers, such as 200-nm liposomes, can also be delivered into cells by TAT peptide attached to the liposome surface. Liposomes were fluorescently labeled with membranotropic rhodamine-phosphatidylethanolamine or by entrapping FITC-dextran. Incubation of fluorescent TAT liposomes with mouse Lewis lung carcinoma cells, human breast tumor BT20 cells, and rat cardiac myocyte H9C2 results in intracellular localization of certain liposomes. Steric hindrances for TAT peptide x cell interaction (attachment of TAT directly to the liposome surface without spacer or the presence of a high MW polyethylene glycol on the liposome surface) abolish liposome internalization, evidencing the importance of direct contact of TAT peptide with the cell surface. Low temperature or metabolic inhibitors, sodium azide or iodoacetamide, have little influence on the translocation of TAT liposomes into cells, confirming the energy-independent character of this process. The approach may have important implications for drug delivery directly into cell cytoplasm.
Figures
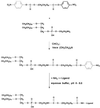
Figure 1
Synthesis of pNP-PEG-PE and its interaction with amino-containing ligand.
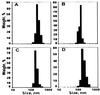
Figure 2
Size distribution patterns for various liposomal preparations used in the study: LIP (A), LIP-NGPE-TAT (B), LIP-PEG(5000) (C), LIP-pNP-PEG(3000) (D).
Figure 3
Schematic pattern of the interaction of TAT peptide-containing liposomes with cell surface: LIP-NGPE-TAT (TAT peptide is located directly on the surface of liposome) (A), LIP-pNP-PEG(3000)-TAT (TAT peptide is coupled via PEG spacer) (B), LIP-PEG(2000)/pNP-PEG(3000)-TAT (TAT peptide is attached to PEGylated liposomes; the length of protecting PEG is smaller that the length of PEG spacer) (C), and LIP-PEG(5000)/pNP-PEG(3000)-TAT (TAT peptide is attached to PEGylated liposomes; the length of protecting PEG is bigger that the length of PEG spacer) (D). 1, phospholipid constituting liposomes; 2, PE anchor for TAT peptide or TAT peptide-bearing PEG spacer; 3, TAT peptide; 4, PEG chains of variable length; and 5, hypothetical sites of TAT peptide interaction with the cell surface.
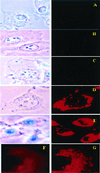
Figure 4
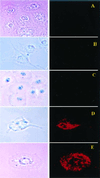
Bright field (Left) and fluorescent (Right) microscopy of cell cultures treated with different preparations of Rh-PE labeled liposomes (37°C; ×600): H9C2 and LIP (A), BT20 and LIP-pNP-PEG(3000) (B), BT20 and LIP-NGPE-TAT (C), BT20 and LIP-pNP-PEG(3000)-TAT (D), and LLC and LIP-pNP-PEG(3000)-TAT (E). (Blue spots seen in pictures under light microscopy are because of variations in light intensity, camera capture settings, and processing.) (F and G) Images represent the results of confocal microscopy of BT20 treated with LIP-pNP-PEG(3000)-TAT at 37°C: single cell viewed from the top cell surface (F); single cell viewed at the mid-point of the cell thickness (≈0.8-μm distance) (G).
Figure 5
Bright field (Left) and fluorescent (Right) microscopy of cell cultures treated with Rh-PE-liposomes (37°C; ×400): BT20 and LIP-PEG(2000) (A), H9C2 and LIP-PEG(5000)/pNP-PEG(3000)-TAT (B), BT20 and LIP-PEG(5000)/pNP-PEG(3000)-TAT (C), H9C2 and LIP-PEG(2000)/pNP-PEG(3000)-TAT (D), and BT20 and LIP-PEG(2000)/pNP-PEG(3000)-TAT (E).
Figure 6
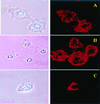
Bright field (Left) and fluorescent (Right) microscopy of viable cells treated with Rh-PE-liposomes in the presence of metabolic inhibitors (×400): BT20 and LIP-pNP-PEG(3000)-TAT, 4°C (A); BT20 and LIP-pNP-PEG(3000)-TAT, 0.1% sodium azide, 37°C (B); and LLC and LIP-pNP-PEG(3000)-TAT, 1% IAA, 37°C (C).
Figure 7
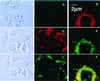
Bright field (Left) and fluorescent (Center) microscopy of H9C2 treated with free FITC-dextran and FITC-dextran-loaded Rh-PE labeled liposomes (37°C; ×400): H9C2 and free FITC-dextran (A); H9C2 and LIP-pNP-PEG(3000)-TAT/FITC-dextran, TRITC filter (B); and H9C2 and LIP-pNP-PEG(3000)-TAT/FITC-dextran, FITC filter (C). (Right) Images represent the results of confocal microscopy of BT20 treated with FITC-dextran-loaded Rh-PE liposomes (37°C; pseudocolor enhancement): single cell under FITC filter (D); single cell under Rh filter (E); and composite of superimposed layers from D and E (F).
Similar articles
- TAT peptide-modified liposomes for intracellular delivery of drugs and DNA.
Torchilin VP. Torchilin VP. Cell Mol Biol Lett. 2002;7(2):265-7. Cell Mol Biol Lett. 2002. PMID: 12097943 No abstract available. - OVCAR-3 cells internalize TAT-peptide modified liposomes by endocytosis.
Fretz MM, Koning GA, Mastrobattista E, Jiskoot W, Storm G. Fretz MM, et al. Biochim Biophys Acta. 2004 Oct 11;1665(1-2):48-56. doi: 10.1016/j.bbamem.2004.06.022. Biochim Biophys Acta. 2004. PMID: 15471570 - TAT-liposomes: a novel intracellular drug carrier.
Torchilin VP, Levchenko TS. Torchilin VP, et al. Curr Protein Pept Sci. 2003 Apr;4(2):133-40. doi: 10.2174/1389203033487298. Curr Protein Pept Sci. 2003. PMID: 12678852 Review. - Endosomolytic activity of cationic liposomes enhances the delivery of human immunodeficiency virus-1 trans-activator protein (TAT) to mammalian cells.
Huang L, Farhood H, Serbina N, Teepe AG, Barsoum J. Huang L, et al. Biochem Biophys Res Commun. 1995 Dec 26;217(3):761-8. doi: 10.1006/bbrc.1995.2838. Biochem Biophys Res Commun. 1995. PMID: 8554596 - Cellular uptake [correction of utake] of the Tat peptide: an endocytosis mechanism following ionic interactions.
Vives E. Vives E. J Mol Recognit. 2003 Sep-Oct;16(5):265-71. doi: 10.1002/jmr.636. J Mol Recognit. 2003. PMID: 14523939 Review.
Cited by
- Cytosolic delivery of nucleic acids: The case of ionizable lipid nanoparticles.
Schlich M, Palomba R, Costabile G, Mizrahy S, Pannuzzo M, Peer D, Decuzzi P. Schlich M, et al. Bioeng Transl Med. 2021 Mar 20;6(2):e10213. doi: 10.1002/btm2.10213. eCollection 2021 May. Bioeng Transl Med. 2021. PMID: 33786376 Free PMC article. Review. - Drug Delivery via Cell Membrane Fusion Using Lipopeptide Modified Liposomes.
Yang J, Bahreman A, Daudey G, Bussmann J, Olsthoorn RC, Kros A. Yang J, et al. ACS Cent Sci. 2016 Sep 28;2(9):621-630. doi: 10.1021/acscentsci.6b00172. Epub 2016 Aug 22. ACS Cent Sci. 2016. PMID: 27725960 Free PMC article. - A review of therapeutic prospects of non-viral gene therapy in the retinal pigment epithelium.
Koirala A, Conley SM, Naash MI. Koirala A, et al. Biomaterials. 2013 Sep;34(29):7158-67. doi: 10.1016/j.biomaterials.2013.06.002. Epub 2013 Jun 22. Biomaterials. 2013. PMID: 23796578 Free PMC article. Review. - Enhanced Fluorescent Protein Activity in Polymer Scaffold-Stabilized Phospholipid Nanoshells Using Neutral Redox Initiator Polymerization Conditions.
Ghosh S, Wang X, Wang J, Nguyen PD, Janczak CM, Aspinwall CA. Ghosh S, et al. ACS Omega. 2018 Nov 30;3(11):15890-15899. doi: 10.1021/acsomega.8b01661. Epub 2018 Nov 26. ACS Omega. 2018. PMID: 30533583 Free PMC article. - Codelivery of a cytotoxin and photosensitiser via a liposomal nanocarrier: a novel strategy for light-triggered cytosolic release.
Yaghini E , Dondi R , Edler KJ , Loizidou M , MacRobert AJ , Eggleston IM . Yaghini E , et al. Nanoscale. 2018 Nov 8;10(43):20366-20376. doi: 10.1039/c8nr04048f. Nanoscale. 2018. PMID: 30376028 Free PMC article.
References
- Green M, Loewenstein P M. Cell. 1988;55:1179–1188. - PubMed
- Frankel A D, Pabo C O. Cell. 1988;55:1189–1193. - PubMed
- Vocero-Akbani A, Lissy N A, Dowdy S F. Methods Enzymol. 2000;322:508–521. - PubMed
- Verhoef K, Koken S E C, Berkhout B. Anal Biochem. 1993;210:210–214. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources