Novel topology of BfpE, a cytoplasmic membrane protein required for type IV fimbrial biogenesis in enteropathogenic Escherichia coli - PubMed (original) (raw)
Novel topology of BfpE, a cytoplasmic membrane protein required for type IV fimbrial biogenesis in enteropathogenic Escherichia coli
T E Blank et al. J Bacteriol. 2001 Aug.
Abstract
Enteropathogenic Escherichia coli (EPEC) produces the bundle-forming pilus (BFP), a type IV fimbria that has been implicated in virulence, autoaggregation, and localized adherence to epithelial cells. The bfpE gene is one of a cluster of bfp genes previously shown to encode functions that direct BFP biosynthesis. Here, we show that an EPEC strain carrying a nonpolar mutation in bfpE fails to autoaggregate, adhere to HEp-2 cells, or form BFP, thereby demonstrating that BfpE is required for BFP biogenesis. BfpE is a cytoplasmic membrane protein of the GspF family. To determine the membrane topology of BfpE, we fused bfpE derivatives containing 3' truncations and/or internal deletions to alkaline phosphatase and/or beta-galactosidase reporter genes, whose products are active only when localized to the periplasm or cytoplasm, respectively. In addition, we constructed BfpE sandwich fusions using a dual alkaline phosphatase/beta-galactosidase reporter cassette and analyzed BfpE deletion derivatives by sucrose density flotation gradient fractionation. The data from these analyses support a topology in which BfpE contains four hydrophobic transmembrane (TM) segments, a large cytoplasmic segment at its N terminus, and a large periplasmic segment near its C terminus. This topology is dramatically different from that of OutF, another member of the GspF family, which has three TM segments and is predominantly cytoplasmic. These findings provide a structural basis for predicting protein-protein interactions required for assembly of the BFP biogenesis machinery.
Figures
FIG. 1
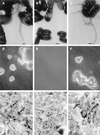
Assay of BFP formation, autoaggregation, and localized adherence by EPEC. Shown are the wild-type EPEC strain E2348/69 (A, D, and G) and the isogenic bfpE mutant UMD934 bearing either the vector pTrcphoA (B, E, and H) or plasmid pTEB41 carrying the bfpE gene (C, F, and I). The top row (A to C) displays transmission electron micrographs of EPEC cultured in DMEM for 6 h. Magnifications, ×20,000 (A) and ×12,000 (B and C). Bar, 200 nm (A) or 500 nm (B and C). BFP can be seen in panels A and C. The center row (D to F) displays phase-contrast micrographs (magnifications, approximately ×460 for panels D and F and ×580 for panel E) of EPEC cultured in DMEM for 7 h and then examined in hanging drop slides. EPEC aggregates are seen in panels D and F. The bottom row (G to I) displays phase-contrast micrographs (magnification, ×630) of HEp-2 cells incubated for 3 hours with EPEC, washed, and fixed. Adherent EPEC microcolonies are seen in panels G and I. Arrows point to two of the microcolonies in each panel.
FIG. 2
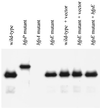
Examination of bundlin expression and processing by EPEC. Whole-cell extracts were prepared from EPEC strains (left to right) E2348/69, UMD932, UMD901, UMD934, E2348/69 (pTrcphoA), UMD934 (pTrcphoA), and UMD934 (pTEB41) after growth in DMEM/F-12 for 6 h. Extracts were separated by SDS-PAGE using a 15% polyacrylamide gel. A monoclonal antibody was used to detect bundlin.
FIG. 3
Expression of BfpE-LacZ fusion proteins. Whole-cell extracts were prepared from plasmid-bearing derivatives of E. coli CC118 and separated by SDS-PAGE on a 6% polyacrylamide gel. The fusion proteins were detected with an anti-β-galactosidase antibody. The positions of molecular mass markers are displayed to the left of the blot. The first three lanes display samples from strains carrying control plasmids pTEB65 (no LacZ), pTrclacZ (no BfpE), and pTEB42 (substrate for exonuclease III digestion). The remaining lanes display samples from strains carrying plasmids with bfpE′::′lacZ fusion genes. The number of the terminal amino acid in the BfpE portion of the fusion protein is noted above each lane. The arrow to the right of the blot indicates a prominent degradation product of many of the fusions that is similar in size to β-galactosidase (LacZ).
FIG. 4
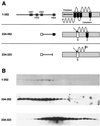
Activities of BfpE-LacZ and BfpE-PhoA fusion proteins. In the diagrams to the left, the shaded bars represent the extent of BfpE included in each fusion protein and the black boxes represent potential transmembrane segments. Black lines and triangles indicate deleted portions of BfpE. Alkaline phosphatase or β-galactosidase enzyme assays were performed on permeabilized cultures of E. coli CC118 carrying fusion plasmids. The bar graph data represent the mean and standard error values (in units of enzyme activity) for four enzyme activity determinations from a single set of permeabilized cells per sample. Similar results were obtained in repeated experiments. Each datum point corresponds to the fusion depicted to the left of it. Four points are absent from the LacZ data (indicated by asterisks) because a plasmid expressing the fusion was not constructed (residue 149) or a fusion protein was not detected by immunoblotting (residues 157, 249, and 352).
FIG. 5
Expression of BfpE-PhoA fusion proteins. Whole-cell extracts were prepared from plasmid-bearing derivatives of E. coli CC118 and separated by SDS-PAGE on a 6.5% polyacrylamide gel. The fusion proteins were detected with an anti-PhoA antibody. The positions of molecular mass markers are displayed to the left of the blot. The first three lanes display samples from strains carrying control plasmids pTrcphoA (no BfpE), pTEB41, and pTEB65 (no PhoA). The remaining lanes display samples from strains carrying plasmids with bfpE′::′phoA fusion genes. The number of the terminal amino acid in the BfpE portion of the fusion protein is noted above each lane. The arrow to the right of the blot indicates a prominent degradation product of many of the fusions that is similar in size to alkaline phosphatase (PhoA).
FIG. 6
Analyses of BfpE–dual-reporter sandwich fusion proteins. The labels indicate restriction sites in the bfpE gene into which a dual-reporter (′_phoA-lacZ_α) cassette was inserted. (A) Activities of sandwich fusion proteins. The BfpE protein is depicted as in Fig. 4, with the arrows indicating the approximate point at which the dual reporter is inserted into the protein. Alkaline phosphatase or β-galactosidase enzyme assays were performed on cultures of E. coli TG1 carrying sandwich fusion plasmids. The bar graph data represent the mean and standard error values for three separate experiments, with four (alkaline phosphatase) or three (β-galactosidase) enzyme activity determinations per experiment. (B) Expression of sandwich fusion proteins. Whole-cell extracts were prepared from the strains described above and separated by SDS-PAGE on a 6% polyacrylamide gel. The fusion proteins were detected with an anti-PhoA antibody. The first three lanes display samples from strains carrying control plasmids, while the remaining lanes display samples from strains carrying sandwich fusions.
FIG. 7
Sucrose density flotation gradient fractionation of three epitope-tagged BfpE derivatives. (A) Representations of epitope-tagged BfpE constructs. Each construct carries a C-terminal _myc_-His double epitope tag (not shown). For each construct, the number in the left column indicates the range of BfpE amino acids that are present. The middle column shows a linear representation of each protein. The right column displays the expected topology of each protein. Black boxes and cylinders indicate TM segments, while white boxes and cylinders indicate an exogenous N-terminal signal sequence, whose presumed removal is indicated by an arrow. (B) Lysates of TOP10 E. coli carrying plasmids producing the constructs shown in panel A were fractionated on sucrose density flotation gradients by centrifugation. Fractions were collected from the top (least dense portion) of the gradient, separated by SDS-PAGE on a 15% polyacrylamide gel, and subjected to immunoblotting using an antiserum recognizing the epitope tag. Fractions progress from the least dense on the left to the most dense on the right.
FIG. 8
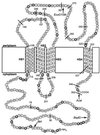
Proposed topology of the BfpE protein. The amino acids composing BfpE are represented by circles. These are displayed in a manner that specifies their arrangement in the E. coli cytoplasmic membrane. HS1 through HS4 denote TM segments. Positively charged amino acids (arginines and lysines) that may be topology determinants are shaded. C-terminal fusion protein junctions are indicated by a line and the number of the terminal BfpE residue in the fusion. The insertion sites for the dual reporter in sandwich fusions are indicated by a restriction enzyme name.
Similar articles
- The inner membrane subassembly of the enteropathogenic Escherichia coli bundle-forming pilus machine.
Crowther LJ, Anantha RP, Donnenberg MS. Crowther LJ, et al. Mol Microbiol. 2004 Apr;52(1):67-79. doi: 10.1111/j.1365-2958.2003.03963.x. Mol Microbiol. 2004. PMID: 15049811 - Structure of an essential type IV pilus biogenesis protein provides insights into pilus and type II secretion systems.
Yamagata A, Milgotina E, Scanlon K, Craig L, Tainer JA, Donnenberg MS. Yamagata A, et al. J Mol Biol. 2012 May 25;419(1-2):110-24. doi: 10.1016/j.jmb.2012.02.041. Epub 2012 Mar 1. J Mol Biol. 2012. PMID: 22387466 Free PMC article. - BfpL is essential for type IV bundle-forming pilus biogenesis and interacts with the periplasmic face of BfpC.
De Masi L, Szmacinski H, Schreiber W, Donnenberg MS. De Masi L, et al. Microbiology (Reading). 2012 Oct;158(Pt 10):2515-2526. doi: 10.1099/mic.0.060889-0. Epub 2012 Jul 26. Microbiology (Reading). 2012. PMID: 22837303 Free PMC article. - Effects of bfp mutations on biogenesis of functional enteropathogenic Escherichia coli type IV pili.
Anantha RP, Stone KD, Donnenberg MS. Anantha RP, et al. J Bacteriol. 2000 May;182(9):2498-506. doi: 10.1128/JB.182.9.2498-2506.2000. J Bacteriol. 2000. PMID: 10762251 Free PMC article.
Cited by
- Purification and three-dimensional electron microscopy structure of the Neisseria meningitidis type IV pilus biogenesis protein PilG.
Collins RF, Saleem M, Derrick JP. Collins RF, et al. J Bacteriol. 2007 Sep;189(17):6389-96. doi: 10.1128/JB.00648-07. Epub 2007 Jul 6. J Bacteriol. 2007. PMID: 17616599 Free PMC article. - The Cpx envelope stress response both facilitates and inhibits elaboration of the enteropathogenic Escherichia coli bundle-forming pilus.
Vogt SL, Nevesinjac AZ, Humphries RM, Donnenberg MS, Armstrong GD, Raivio TL. Vogt SL, et al. Mol Microbiol. 2010 Jun 1;76(5):1095-110. doi: 10.1111/j.1365-2958.2010.07145.x. Epub 2010 Apr 14. Mol Microbiol. 2010. PMID: 20444097 Free PMC article. - The Cpx envelope stress response affects expression of the type IV bundle-forming pili of enteropathogenic Escherichia coli.
Nevesinjac AZ, Raivio TL. Nevesinjac AZ, et al. J Bacteriol. 2005 Jan;187(2):672-86. doi: 10.1128/JB.187.2.672-686.2005. J Bacteriol. 2005. PMID: 15629938 Free PMC article. - Pseudomonas aeruginosa β-lactamase induction requires two permeases, AmpG and AmpP.
Kong KF, Aguila A, Schneper L, Mathee K. Kong KF, et al. BMC Microbiol. 2010 Dec 30;10:328. doi: 10.1186/1471-2180-10-328. BMC Microbiol. 2010. PMID: 21192796 Free PMC article. - Crystal Structure of the Minor Pilin CofB, the Initiator of CFA/III Pilus Assembly in Enterotoxigenic Escherichia coli.
Kolappan S, Ng D, Yang G, Harn T, Craig L. Kolappan S, et al. J Biol Chem. 2015 Oct 23;290(43):25805-18. doi: 10.1074/jbc.M115.676106. Epub 2015 Aug 31. J Biol Chem. 2015. PMID: 26324721 Free PMC article.
References
- Alexeyev M F, Winkler H H. Membrane topology of the Rickettsia prowazekii ATP/ADP translocase revealed by novel dual pho-lac reporters. J Mol Biol. 1999;285:1503–1513. - PubMed
- Amann E, Ochs B, Abel K J. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene. 1988;69:301–315. - PubMed
- Baldini M M, Kaper J B, Levine M M, Candy D C, Moon H W. Plasmid-mediated adhesion in enteropathogenic Escherichia coli. J Pediatr Gastroenterol Nutr. 1983;2:534–538. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- F32 AI010191/AI/NIAID NIH HHS/United States
- R01 AI037606/AI/NIAID NIH HHS/United States
- F32 AI-10191/AI/NIAID NIH HHS/United States
- R01 AI-37606/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources