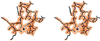Intertwined structure of the DNA-binding domain of intron endonuclease I-TevI with its substrate - PubMed (original) (raw)
Intertwined structure of the DNA-binding domain of intron endonuclease I-TevI with its substrate
P Van Roey et al. EMBO J. 2001.
Abstract
I-TevI is a site-specific, sequence-tolerant intron endonuclease. The crystal structure of the DNA-binding domain of I-TevI complexed with the 20 bp primary binding region of its DNA target reveals an unusually extended structure composed of three subdomains: a Zn finger, an elongated segment containing a minor groove-binding alpha-helix, and a helix-turn-helix. The protein wraps around the DNA, mostly following the minor groove, contacting the phosphate backbone along the full length of the duplex. Surprisingly, while the minor groove-binding helix and the helix-turn- helix subdomain make hydrophobic contacts, the few base-specific hydrogen bonds occur in segments that lack secondary structure and flank the intron insertion site. The multiple base-specific interactions over a long segment of the substrate are consistent with the observed high site specificity in spite of sequence tolerance, while the modular composition of the domain is pertinent to the evolution of homing endonucleases.
Figures
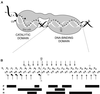
Fig. 1. I-_Tev_I–DNA interaction. (A) Cartoon of the two-domain structure of I-_Tev_I with its DNA homing site. (B) Protein–DNA contacts. The sequence of the DNA fragment used in the crystal structure determination with interaction sites as identified from ethylation and methylation protection assays indicated as arrows and circles (open, weak; closed, strong), respectively. Bars below indicate the DNA locations of the three types of protein–DNA contacts in the structure, showing the consistency with the biochemical data. P and H correspond to regions involved in phosphate–backbone and hydrophobic contacts, respectively, while B refers to base-specific hydrogen-bonding contacts. IS and CS indicate the insertion and cleavage sites, respectively.
Fig. 2. Electron density map for the Zn finger subdomain. Stereodiagram showing the final (2_F_o – _F_c) map for residues 149–168, contoured at the 1.5σ level. The Zn ion is shown as a blue sphere in the lower center of the view and the 10 residue loop oriented towards the top of the figure. The figure was prepared using SETOR (Evans, 1993).
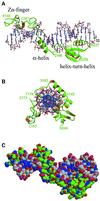
Fig. 3. Three-dimensional structure of the complex of the DNA-binding domain of I-_Tev_I with its substrate. The complex is shown (A) perpendicular to the DNA axis and (B) along the DNA axis. Distortions to the DNA are limited to widening of the minor groove. (C) Space-filling model of the structure of the complex, showing the continuous tight association between the two molecules. Protein and DNA carbon atoms are colored green and gray, respectively. Figures 3 and 4 were prepared with Molscript (Kraulis, 1991) and Raster3D (Merritt and Bacon, 1997).
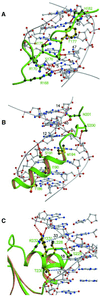
Fig. 4. Individual I-_Tev_I–DNA contact regions. (A) Elongated segment between the Zn finger and the minor groove-binding helix. The protein lacks secondary structure but residues 170–180 form a twisted structure that widens the minor groove. Base-specific hydrogen-bonding contacts (red dotted lines) throughout this segment are interrupted by the hydrophobic insertion of the phenyl ring of Phe177 (yellow). (B) The minor groove-binding α-helix. Only one hydrogen-bonding contact is seen in the helix (Ser191 to the phosphate backbone), and the surface close to the DNA consists of three hydrophobic residues. The section between the helix and the helix–turn–helix subdomain includes the remaining base-specific hydrogen-bonding contacts, Asn201 to GUA40 and CYT14. (C) The helix–turn–helix subdomain inserts its second helix into the major groove. Several of the residues of this helix make hydrogen-bonding contacts to the phosphate backbone (red dotted lines), but the surface of the helix adjacent to the DNA is mostly hydrophobic and matches the hydrophobic surface of the DNA, which presents the C5-methyl groups of thymidines 16, 17, 18, 33 and 34. The closest hydrophobic contacts are shown as blue dotted lines.
Similar articles
- Zinc finger as distance determinant in the flexible linker of intron endonuclease I-TevI.
Dean AB, Stanger MJ, Dansereau JT, Van Roey P, Derbyshire V, Belfort M. Dean AB, et al. Proc Natl Acad Sci U S A. 2002 Jun 25;99(13):8554-61. doi: 10.1073/pnas.082253699. Epub 2002 Jun 19. Proc Natl Acad Sci U S A. 2002. PMID: 12077294 Free PMC article. - The td intron endonuclease I-TevI makes extensive sequence-tolerant contacts across the minor groove of its DNA target.
Bryk M, Quirk SM, Mueller JE, Loizos N, Lawrence C, Belfort M. Bryk M, et al. EMBO J. 1993 May;12(5):2141-9. doi: 10.1002/j.1460-2075.1993.tb05862.x. EMBO J. 1993. PMID: 8491202 Free PMC article. - Distance determination by GIY-YIG intron endonucleases: discrimination between repression and cleavage functions.
Liu Q, Derbyshire V, Belfort M, Edgell DR. Liu Q, et al. Nucleic Acids Res. 2006 Mar 31;34(6):1755-64. doi: 10.1093/nar/gkl079. Print 2006. Nucleic Acids Res. 2006. PMID: 16582101 Free PMC article. - Homing endonucleases: structural and functional insight into the catalysts of intron/intein mobility.
Chevalier BS, Stoddard BL. Chevalier BS, et al. Nucleic Acids Res. 2001 Sep 15;29(18):3757-74. doi: 10.1093/nar/29.18.3757. Nucleic Acids Res. 2001. PMID: 11557808 Free PMC article. Review. - DNA recognition and structural specificities.
Roy KB. Roy KB. Indian J Biochem Biophys. 1996 Apr;33(2):83-7. Indian J Biochem Biophys. 1996. PMID: 8754617 Review.
Cited by
- Activity, specificity and structure of I-Bth0305I: a representative of a new homing endonuclease family.
Taylor GK, Heiter DF, Pietrokovski S, Stoddard BL. Taylor GK, et al. Nucleic Acids Res. 2011 Dec;39(22):9705-19. doi: 10.1093/nar/gkr669. Epub 2011 Sep 2. Nucleic Acids Res. 2011. PMID: 21890897 Free PMC article. - Intein-mediated purification of cytotoxic endonuclease I-TevI by insertional inactivation and pH-controllable splicing.
Wu W, Wood DW, Belfort G, Derbyshire V, Belfort M. Wu W, et al. Nucleic Acids Res. 2002 Nov 15;30(22):4864-71. doi: 10.1093/nar/gkf621. Nucleic Acids Res. 2002. PMID: 12433989 Free PMC article. - Mycobacterium tuberculosis RecA intein, a LAGLIDADG homing endonuclease, displays Mn(2+) and DNA-dependent ATPase activity.
Guhan N, Muniyappa K. Guhan N, et al. Nucleic Acids Res. 2003 Jul 15;31(14):4184-91. doi: 10.1093/nar/gkg475. Nucleic Acids Res. 2003. PMID: 12853636 Free PMC article. - Functional Validation of Different Alternative Splicing Variants of the Chrysanthemum lavandulifolium ClNUM1 Gene in Tobacco.
Zhang W, Wang H, Guo Y, Hao X, Li Y, He W, Zhao X, Cai S, Song X. Zhang W, et al. Curr Issues Mol Biol. 2024 May 25;46(6):5242-5256. doi: 10.3390/cimb46060314. Curr Issues Mol Biol. 2024. PMID: 38920986 Free PMC article. - Structural classification of zinc fingers: survey and summary.
Krishna SS, Majumdar I, Grishin NV. Krishna SS, et al. Nucleic Acids Res. 2003 Jan 15;31(2):532-50. doi: 10.1093/nar/gkg161. Nucleic Acids Res. 2003. PMID: 12527760 Free PMC article.
References
- Belfort M., Derbyshire,V., Parker,M.M., Cousineau,B. and Lambowitz,A.M. (2001) Mobile introns: pathways and proteins. In Craig,N., Craige,R., Gellert,M. and Lambowitz,A. (eds), Mobile DNA II. ASM Press, in press.
- Brünger A.T. et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. - PubMed
- Bryk M., Belisle,M., Mueller,J.E. and Belfort,M. (1995) Selection of a remote cleavage site by I-TevI, the td intron-encoded endonuclease. J. Mol. Biol., 247, 197–201. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM44844/GM/NIGMS NIH HHS/United States
- R37 GM039422/GM/NIGMS NIH HHS/United States
- GM39422/GM/NIGMS NIH HHS/United States
- R01 GM039422/GM/NIGMS NIH HHS/United States
- GM56966/GM/NIGMS NIH HHS/United States
- R01 GM044844/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases