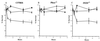Cu,Zn superoxide dismutase of Mycobacterium tuberculosis contributes to survival in activated macrophages that are generating an oxidative burst - PubMed (original) (raw)
Cu,Zn superoxide dismutase of Mycobacterium tuberculosis contributes to survival in activated macrophages that are generating an oxidative burst
D L Piddington et al. Infect Immun. 2001 Aug.
Abstract
Macrophages produce reactive oxygen species and reactive nitrogen species that have potent antimicrobial activity. Resistance to killing by macrophages is critical to the virulence of Mycobacterium tuberculosis. M. tuberculosis has two genes encoding superoxide dismutase proteins, sodA and sodC. SodC is a Cu,Zn superoxide dismutase responsible for only a minor portion of the superoxide dismutase activity of M. tuberculosis. However, SodC has a lipoprotein binding motif, which suggests that it may be anchored in the membrane to protect M. tuberculosis from reactive oxygen intermediates at the bacterial surface. To examine the role of the Cu,Zn superoxide dismutase in protecting M. tuberculosis from the toxic effects of exogenously generated reactive oxygen species, we constructed a null mutation in the sodC gene. In this report, we show that the M. tuberculosis sodC mutant is readily killed by superoxide generated externally, while the isogenic parental M. tuberculosis is unaffected under these conditions. Furthermore, the sodC mutant has enhanced susceptibility to killing by gamma interferon (IFN-gamma)-activated murine peritoneal macrophages producing oxidative burst products but is unaffected by macrophages not activated by IFN-gamma or by macrophages from respiratory burst-deficient mice. These observations establish that the Cu,Zn superoxide dismutase contributes to the resistance of M. tuberculosis against oxidative burst products generated by activated macrophages.
Figures
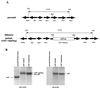
FIG. 1
(A) Restriction map of the sodC region in parental and sodC mutant strains of M. tuberculosis. (B) Southern blot of chromosomal DNAs from the parental, mutant, and complemented strains. DNA was digested with _Eco_RV and probed with DNA containing the gene for sodC or for aph. The _sodC-_hybridizing fragment in mutant Mtb1612 is larger than the unmutated fragment due to insertion of the drug resistance cassette. This fragment also hybridizes with the aph probe, confirming that Mtb1612 contains the kanamycin resistance gene that produced the mutation.
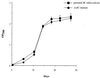
FIG. 2
Growth of the sodC mutant Mtb1612 and parental M. tuberculosis Erdman in 7H9 broth. Growth was monitored by reading the OD580 and is expressed as the mean and standard error of the mean for triplicate samples.
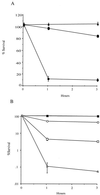
FIG. 3
(A) Survival of the sodC mutant Mtb1612 (circles) and the _sodC_-complemented strain Mtb1623 (triangles) was compared to survival of parental M. tuberculosis (squares) using hypoxanthine/xanthine oxidase to generate superoxide. The number of surviving bacteria was determined at 0, 1, and 3 h after exposure to superoxide by plating dilutions of the bacteria on 7H10 plates. The means from triplicate tubes were calculated, and the data are expressed as percentages of the time zero value. Results of a representative assay from six experiments are shown. (B) Survival of the sodC mutant Mtb1612 (open symbols) was compared to that of parental M. tuberculosis (closed symbols) using hypoxanthine/xanthine oxidase and SPER/NO (Alexis Biochemicals) to generate superoxide (squares), nitric oxide (circles), or a combination of both (triangles). The number of surviving bacteria was determined at 0, 1, and 3 h after exposure to the compounds by plating dilutions on 7H10 medium. The means from triplicate tubes were calculated, and the data are expressed as percentages of the time zero value. Results of a representative assay from three experiments are shown.
FIG. 4
Survival of parental M. tuberculosis (squares), the sodC mutant Mtb1612 (circles), or the _sodC_-complemented strain Mtb1623 (triangles) in peritoneal macrophages from C57BL/6 mice (A), gp91_phox_−/− mice (B), or iNOS−/− mice (C). Macrophages were activated with IFN-γ (100 U/ml) overnight and were then infected with bacteria at a multiplicity of infection of 10:1. The number of surviving bacteria was determined by plating dilutions of the macrophage lysate on 7H10 plates. The data are expressed as the mean and standard error of the mean from triplicate wells (C57BL/6 and iNOS−/−) or quadruplicate wells (gp91_phox_−/−) at each time point.
FIG. 5
Southern blot of chromosomal DNAs from 15 mycobacterial species. DNA was digested with _Not_I and probed with the sodC gene from M. tuberculosis. Lane 1, M. tuberculosis; lane 2, M. africanum; lane 3, M. bovis; lane 4, M. microti; lane 5, M. bovis BCG; lane 6, M. fortuitum; lane 7, M. chelonae; lane 8, M. smegmatis; lane 9, M. avium; lane 10, M. intracellulare; lane 11, M. gordonii; lane 12, M. marinum; lane 13, M. scrofulaceum; lane 14, M. kansasii; and lane 15, M. xenopi.
Similar articles
- The Brucella abortus Cu,Zn superoxide dismutase is required for optimal resistance to oxidative killing by murine macrophages and wild-type virulence in experimentally infected mice.
Gee JM, Valderas MW, Kovach ME, Grippe VK, Robertson GT, Ng WL, Richardson JM, Winkler ME, Roop RM 2nd. Gee JM, et al. Infect Immun. 2005 May;73(5):2873-80. doi: 10.1128/IAI.73.5.2873-2880.2005. Infect Immun. 2005. PMID: 15845493 Free PMC article. - Periplasmic superoxide dismutase protects Salmonella from products of phagocyte NADPH-oxidase and nitric oxide synthase.
De Groote MA, Ochsner UA, Shiloh MU, Nathan C, McCord JM, Dinauer MC, Libby SJ, Vazquez-Torres A, Xu Y, Fang FC. De Groote MA, et al. Proc Natl Acad Sci U S A. 1997 Dec 9;94(25):13997-4001. doi: 10.1073/pnas.94.25.13997. Proc Natl Acad Sci U S A. 1997. PMID: 9391141 Free PMC article. - Characterization of SodC, a periplasmic superoxide dismutase from Burkholderia cenocepacia.
Keith KE, Valvano MA. Keith KE, et al. Infect Immun. 2007 May;75(5):2451-60. doi: 10.1128/IAI.01556-06. Epub 2007 Feb 26. Infect Immun. 2007. PMID: 17325048 Free PMC article. - Role of prokaryotic Cu,Zn superoxide dismutase in pathogenesis.
Battistoni A. Battistoni A. Biochem Soc Trans. 2003 Dec;31(Pt 6):1326-9. doi: 10.1042/bst0311326. Biochem Soc Trans. 2003. PMID: 14641055 Review. - NADPH oxidase, Nramp1 and nitric oxide synthase 2 in the host antimicrobial response.
Karupiah G, Hunt NH, King NJ, Chaudhri G. Karupiah G, et al. Rev Immunogenet. 2000;2(3):387-415. Rev Immunogenet. 2000. PMID: 11256747 Review.
Cited by
- Long-range charge transfer mechanism of the III2IV2 mycobacterial supercomplex.
Riepl D, Gamiz-Hernandez AP, Kovalova T, Król SM, Mader SL, Sjöstrand D, Högbom M, Brzezinski P, Kaila VRI. Riepl D, et al. Nat Commun. 2024 Jun 20;15(1):5276. doi: 10.1038/s41467-024-49628-9. Nat Commun. 2024. PMID: 38902248 Free PMC article. - Live Attenuated Vaccines against Tuberculosis: Targeting the Disruption of Genes Encoding the Secretory Proteins of Mycobacteria.
Veerapandian R, Gadad SS, Jagannath C, Dhandayuthapani S. Veerapandian R, et al. Vaccines (Basel). 2024 May 12;12(5):530. doi: 10.3390/vaccines12050530. Vaccines (Basel). 2024. PMID: 38793781 Free PMC article. Review. - Lipid Peroxidation and Type I Interferon Coupling Fuels Pathogenic Macrophage Activation Causing Tuberculosis Susceptibility.
Yabaji SM, Zhernovkov V, Araveti PB, Lata S, Rukhlenko OS, Al Abdullatif S, Vanvalkenburg A, Alekseev Y, Ma Q, Dayama G, Lau NC, Johnson WE, Bishai WR, Crossland NA, Campbell JD, Kholodenko BN, Gimelbrant AA, Kobzik L, Kramnik I. Yabaji SM, et al. bioRxiv [Preprint]. 2024 Sep 25:2024.03.05.583602. doi: 10.1101/2024.03.05.583602. bioRxiv. 2024. PMID: 38496444 Free PMC article. Preprint. - The Benefits of Toxicity: M. smegmatis VapBC TA Module Is Induced by Tetracycline Exposure and Promotes Survival.
Zamakhaev M, Bespyatykh J, Goncharenko A, Shumkov M. Zamakhaev M, et al. Microorganisms. 2023 Nov 26;11(12):2863. doi: 10.3390/microorganisms11122863. Microorganisms. 2023. PMID: 38138007 Free PMC article. - Targeted Mutagenesis of Mycobacterium Strains by Homologous Recombination.
Song S, Su Z. Song S, et al. Methods Mol Biol. 2023;2704:85-96. doi: 10.1007/978-1-0716-3385-4_5. Methods Mol Biol. 2023. PMID: 37642839
References
- Absolom D. Basic methods for the study of phagocytosis. Methods Enzymol. 1986;132:95–180. - PubMed
- Adams L B, Dinauer M, Morgenstern D, Krahenbuhl J. Comparison of the roles of reactive oxygen and nitrogen intermediates in the host response to Mycobacterium tuberculosis using transgenic mice. Tuber Lung Dis. 1997;78:237–246. - PubMed
- Bogdan C, Rollinghoff M, Diefenbach A. Reactive oxygen and reactive nitrogen intermediates in innate and specific immunity. Curr Opin Immunol. 2000;12:64–76. - PubMed
- Bryk R, Griffin P, Nathan C. Peroxynitrite reductase activity of bacterial peroxiredoxins. Nature. 2000;407:211–215. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI040488/AI/NIAID NIH HHS/United States
- AI39557/AI/NIAID NIH HHS/United States
- R01 AI039557/AI/NIAID NIH HHS/United States
- AI44486/AI/NIAID NIH HHS/United States
- R01 AI044486/AI/NIAID NIH HHS/United States
- N01AI40075/AI/NIAID NIH HHS/United States
- AI40488/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous