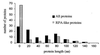Rolling-circle transposons in eukaryotes - PubMed (original) (raw)
Rolling-circle transposons in eukaryotes
V V Kapitonov et al. Proc Natl Acad Sci U S A. 2001.
Abstract
All eukaryotic DNA transposons reported so far belong to a single category of elements transposed by the so-called "cut-and-paste" mechanism. Here, we report a previously unknown category of eukaryotic DNA transposons, Helitron, which transpose by rolling-circle replication. Autonomous Helitrons encode a 5'-to-3' DNA helicase and nuclease/ligase similar to those encoded by known rolling-circle replicons. Helitron-like transposons have conservative 5'-TC and CTRR-3' termini and do not have terminal inverted repeats. They contain 16- to 20-bp hairpins separated by 10--12 nucleotides from the 3'-end and transpose precisely between the 5'-A and T-3', with no modifications of the AT target sites. Together with their multiple diverged nonautonomous descendants, Helitrons constitute approximately 2% of both the Arabidopsis thaliana and Caenorhabditis elegans genomes and also colonize the Oriza sativa genome. Sequence conservation suggests that Helitrons continue to be transposed.
Figures
Figure 1
Reconstruction of the Helitron1 (A),Helitron2 (B), and_Helitron1_CE_ (C) consensus sequences. The consensus sequences are schematically depicted as rectangles. Contiguous copies of Helitrons that we used for reconstruction of the consensus sequences are shown as bold lines beneath the rectangles. Gaps in the lines mark deletions of corresponding regions of the consensus sequences. GenBank accession nos. and sequence coordinates are indicated. Genes and their coordinates in the consensus sequences are indicated above the rectangles. Genes coding for proteins composed of the Rep and helicase domains are shaded in gray. The AT target sites are encircled.
Figure 2
Multiple alignment of helicases encoded by the Helitron1_and Helitron1_CE transposons with a set of eukaryotic and prokaryotic DNA helicases. Domains I–VI that are conservative in DNA helicases from the SF1 superfamily (20) and distances between these domains are indicated. Domain IV/V has not been reported previously. Invariable positions are shaded in black, and those conserved in more than 60% of the sequences are shaded in gray. The following are names of helicases: PIF1 (GenBank protein identification no. 130196, yeast), BACULOVIRUS (7460536, the dsDNA Lymantia dispar_nucleopolyhedrovirus), CHILO (5725645, the dsDNA chilo iridescent virus), TRAA_RHISN (2499024, a Ti-like plasmid from_Rhizobium), TRAI_EC (136208, the F plasmid from_Escherichia coli), EXOV_EC (2507018, the RecD subunit from the E. coli exodeoxyribonuclease V), TRWC (1084124, the R388 conjugative plasmid from E. coli), and HEL_T4 (416895, the dsDNA T4 bacteriophage).
Figure 3
Termini of Helitrons. Conserved 5′ and 3′ termini are in bold capital letters, 3′ terminal hairpins are shaded in gray, and inverted repeats are underlined.
Figure 4
Precise integration of Helitrons into the host AT target sites. Six insertion cases of Helitrons (red) into different transposons (green) are shown separately. Two copies of ATREP1, two copies of Helitrony3_CE, and single copies of Helitrony2_CE and ATREP9 are inserted into copies of the ATTIRX1D, ATHATN1, PAL5A_CE, LTR2_CE, PALTTAA1_CE, and ATREP10 transposons, respectively. The consensus sequences of the elements harboring _Helitron_s are marked by the bold black letters and are described in the _A. thaliana_and C. elegans sections of Repbase Update at
www.girinst.org/Repbase\_Update.html
. Consensus sequences of the corresponding Helitrons are marked in blue. Asterisks, semicolons, and dots indicate identical nucleotide positions, transitions, and transversions, respectively. Only termini of_Helitrons_ are shown. Black, green, and blue numbers show positions in the consensus sequences of the harboring transposons, GenBank, and Helitron consensus sequences, respectively.
Figure 5
Distributions of protein lengths predicted in 100 random DNA sequences. Every random sequence was 61% identical to ATRPA2H, without insertions or deletions. Black marks all proteins predicted by
genscan
; gray marks proteins similar to ATRPA2Hp.
Figure 6
Alignment of the RC motifs in the Helitrons. Following is a list of the RCR initiator-like proteins: SVTS2 (GenBank accession no. AAF18310, the SVTS2 ssDNA spiroplasma plectrovirus); Rep_SC (BAA34784, the pSA1.1 conjugative plasmid from Streptomyces cyaneus); Rep_BB (BAA07788, the pHT926 Bacillus borstelensis cryptic plasmid); Rep_AA (AAC37125, the pVT736–1 RCR plasmid from Actinobacillus actinomycetemcomitans); and Pf3 (AAA88392, the Pf3 ssDNA bacteriophage from Pseudomonas aeruginosa). Color shading shows different physicochemical properties of conserved amino acids (17). Conserved tyrosines, corresponding to the RCR nicking/ligation catalytic center, are marked by dots.
Comment in
- Treasures in the attic: rolling circle transposons discovered in eukaryotic genomes.
Feschotte C, Wessler SR. Feschotte C, et al. Proc Natl Acad Sci U S A. 2001 Jul 31;98(16):8923-4. doi: 10.1073/pnas.171326198. Proc Natl Acad Sci U S A. 2001. PMID: 11481459 Free PMC article. No abstract available.
Similar articles
- Helitrons on a roll: eukaryotic rolling-circle transposons.
Kapitonov VV, Jurka J. Kapitonov VV, et al. Trends Genet. 2007 Oct;23(10):521-9. doi: 10.1016/j.tig.2007.08.004. Epub 2007 Sep 11. Trends Genet. 2007. PMID: 17850916 Review. - Rolling-circle amplification of centromeric Helitrons in plant genomes.
Xiong W, Dooner HK, Du C. Xiong W, et al. Plant J. 2016 Dec;88(6):1038-1045. doi: 10.1111/tpj.13314. Epub 2016 Oct 25. Plant J. 2016. PMID: 27553634 - Spontaneous mutations caused by a Helitron transposon, Hel-It1, in morning glory, Ipomoea tricolor.
Choi JD, Hoshino A, Park KI, Park IS, Iida S. Choi JD, et al. Plant J. 2007 Mar;49(5):924-34. doi: 10.1111/j.1365-313X.2006.03007.x. Epub 2007 Jan 25. Plant J. 2007. PMID: 17257169 - Highly expressed captured genes and cross-kingdom domains present in Helitrons create novel diversity in Pleurotus ostreatus and other fungi.
Castanera R, Pérez G, López L, Sancho R, Santoyo F, Alfaro M, Gabaldón T, Pisabarro AG, Oguiza JA, Ramírez L. Castanera R, et al. BMC Genomics. 2014 Dec 5;15(1):1071. doi: 10.1186/1471-2164-15-1071. BMC Genomics. 2014. PMID: 25480150 Free PMC article. - Helitrons: genomic parasites that generate developmental novelties.
Barro-Trastoy D, Köhler C. Barro-Trastoy D, et al. Trends Genet. 2024 May;40(5):437-448. doi: 10.1016/j.tig.2024.02.002. Epub 2024 Feb 29. Trends Genet. 2024. PMID: 38429198 Review.
Cited by
- Hidden magicians of genome evolution.
Kumar CS, Qureshi SF, Ali A, Satyanarayana ML, Rangaraju A, Venkateshwari A, Nallari P. Kumar CS, et al. Indian J Med Res. 2013 Jun;137(6):1052-60. Indian J Med Res. 2013. PMID: 23852286 Free PMC article. Review. - Transduction of RNA-directed DNA methylation signals to repressive histone marks in Arabidopsis thaliana.
Numa H, Kim JM, Matsui A, Kurihara Y, Morosawa T, Ishida J, Mochizuki Y, Kimura H, Shinozaki K, Toyoda T, Seki M, Yoshikawa M, Habu Y. Numa H, et al. EMBO J. 2010 Jan 20;29(2):352-62. doi: 10.1038/emboj.2009.374. Epub 2009 Dec 10. EMBO J. 2010. PMID: 20010696 Free PMC article. - Fosmid library construction and initial analysis of end sequences in Zhikong scallop (Chlamys farreri).
Zhang L, Bao Z, Cheng J, Li H, Huang X, Wang S, Zhang C, Hu J. Zhang L, et al. Mar Biotechnol (NY). 2007 Sep-Oct;9(5):606-12. doi: 10.1007/s10126-007-9014-4. Epub 2007 Jun 29. Mar Biotechnol (NY). 2007. PMID: 17605073 - Distribution, diversity, evolution, and survival of Helitrons in the maize genome.
Yang L, Bennetzen JL. Yang L, et al. Proc Natl Acad Sci U S A. 2009 Nov 24;106(47):19922-7. doi: 10.1073/pnas.0908008106. Epub 2009 Nov 19. Proc Natl Acad Sci U S A. 2009. PMID: 19926865 Free PMC article. - Effects of recombination rate and gene density on transposable element distributions in Arabidopsis thaliana.
Wright SI, Agrawal N, Bureau TE. Wright SI, et al. Genome Res. 2003 Aug;13(8):1897-903. doi: 10.1101/gr.1281503. Genome Res. 2003. PMID: 12902382 Free PMC article.
References
- Berg D E, Howe M M, editors. Mobile DNA. Washington, DC: Am. Soc. Microbiol.; 1989.
- Fedoroff N V. Ann NY Acad Sci. 1999;870:251–264. - PubMed
- Craig N L. Science. 1995;270:253–254. - PubMed
- Turlan C, Chandler M. Trends Microbiol. 2000;8:268–274. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources