Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene - PubMed (original) (raw)
Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene
L Bartee et al. Genes Dev. 2001.
Abstract
Plants maintain cytosine methylation at CG and non-CG residues to control gene expression and genome stability. In a screen for Arabidopsis mutants that alter methylation and silencing of a densely methylated endogenous reporter gene, we recovered 11 loss-of-function alleles in the CMT3 chromomethylase gene. The cmt3 mutants displayed enhanced expression and reduced methylation of the reporter, particularly at non-CG cytosines. CNG methylation was also reduced at repetitive centromeric sequences. Thus, CMT3 is a key determinant for non-CG methylation. The lack of CMT homologs in animal genomes could account for the observation that in contrast to plants, animals maintain primarily CG methylation.
Figures
Figure 1
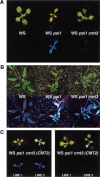
Wassilewskija (WS) pai1C251Y cmt3 plant phenotypes. (A) Two-week-old seedlings of the indicated genotypes grown on agar medium are shown under visible (top) and UV (bottom) light. (B) Four-week-old adult plants of the indicated genotypes grown in soil are shown under visible (top) and UV (bottom) light. (C) Representative 2-week-old T2 generation transgenic seedlings of the indicated genotypes grown on agar medium are shown under visible (top) and UV (bottom) light. The phenotypes of the cmt3G456D allele shown are representative of the phenotypes observed with 10 other cmt3 alleles.
Figure 2
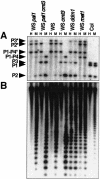
cmt3 mutations confer reduced PAI and CEN methylation. (A) Genomic DNAs prepared from 4-week-old plants of the indicated genotypes were cleaved with either _Hpa_II (H) or _Msp_I (M) and used for Southern blot analysis with a PAI probe (Bender and Fink 1995). (P1–P4) PAI1–PAI4; (P2) PAI2; (P3) PAI3; (P1) Columbia (Col) strain PAI1; asterisks indicate the positions of species methylated at internal _Hpa_II/_Msp_I sites (Bender and Fink 1995; Luff et al. 1999). The Col strain is included as a control for the positions of unmethylated PAI2 and PAI3 species. (B) The blot shown in A was reprobed with a 180-bp CEN repeat probe. The phenotypes of the cmt3G456D allele shown are representative of the phenotypes observed with 10 other cmt3 alleles.
Figure 3
Sequencing of PAI promoter methylation in the cmt3 mutant. Bisulfite genomic methylation sequencing was performed as described (Jeddeloh et al. 1998; Luff et al. 1999) for the top strands of the PAI1 and PAI2 promoters in Wassilewskija (WS) pai1C251Y cmt3G456D DNA. For each region, eight independent molecules were sequenced. Vertical lines indicate positions of cytosines, with the height of each line representing how many sequenced molecules had 5-methyl-cytosine. (Black) CG cytosines; (blue) CNG cytosines; (red) other cytosines. Asterisks indicate sites with no methylation. The black horizontal line indicates the region of PAI identity, and the gray horizontal line indicates flanking upstream heterologous sequence unique to each gene. The exon and intron structures of PAI1 and PAI2 are shown as open boxes and dashed lines, respectively, under each sequence. These structures are based on full-length cDNA sequences for each gene (Melquist et al. 1999).
Figure 4
Positions of mutations in CMT3. The predicted amino acid sequences of Wassilewskiya (WS) CMT3, WS CMT2, and maize ZMET2 are shown aligned along their conserved C-terminal regions. The N termini, upstream of the backslash at the beginning of each sequence, are unrelated. CMT3 introns are indicated by inverted triangles above the sequence. CMT3 missense mutations are indicated above the affected residues. The stop mutation is indicated by an asterisk, and the splice donor and acceptor site mutations are indicated by an x to the left or right, respectively, above the affected introns. Residues identical between proteins are highlighted in boldface. Conserved sequence motifs are indicated under the alignment. GenBank accession nos. are: AF383170 for WS CMT3 and AF383171 for WS CMT2.
Similar articles
- Characterization of non-CG genomic hypomethylation associated with gamma-ray-induced suppression of CMT3 transcription in Arabidopsis thaliana.
Kim JE, Lee MH, Cho EJ, Kim JH, Chung BY, Kim JH. Kim JE, et al. Radiat Res. 2013 Dec;180(6):638-48. doi: 10.1667/RR13394.1. Epub 2013 Nov 26. Radiat Res. 2013. PMID: 24279389 - RNAi, DRD1, and histone methylation actively target developmentally important non-CG DNA methylation in arabidopsis.
Chan SW, Henderson IR, Zhang X, Shah G, Chien JS, Jacobsen SE. Chan SW, et al. PLoS Genet. 2006 Jun 2;2(6):e83. doi: 10.1371/journal.pgen.0020083. PLoS Genet. 2006. PMID: 16741558 Free PMC article. - Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation.
Lindroth AM, Cao X, Jackson JP, Zilberman D, McCallum CM, Henikoff S, Jacobsen SE. Lindroth AM, et al. Science. 2001 Jun 15;292(5524):2077-80. doi: 10.1126/science.1059745. Epub 2001 May 10. Science. 2001. PMID: 11349138 - Silencing of transposons in plant genomes: kick them when they're down.
Zilberman D, Henikoff S. Zilberman D, et al. Genome Biol. 2004;5(12):249. doi: 10.1186/gb-2004-5-12-249. Epub 2004 Nov 16. Genome Biol. 2004. PMID: 15575975 Free PMC article. Review. - Methylation and demethylation of the Arabidopsis genome.
Furner IJ, Matzke M. Furner IJ, et al. Curr Opin Plant Biol. 2011 Apr;14(2):137-41. doi: 10.1016/j.pbi.2010.11.004. Epub 2010 Dec 13. Curr Opin Plant Biol. 2011. PMID: 21159546 Review.
Cited by
- Revealing cis- and trans-regulatory elements underlying nuclear distribution and function of the Arabidopsis histone H2B.8 variant.
Khadka J, Trishla VS, Sannidhi S, Singiri JR, Grandhi R, Pesok A, Novoplansky N, Adler-Agmon Z, Grafi G. Khadka J, et al. BMC Plant Biol. 2024 Aug 28;24(1):811. doi: 10.1186/s12870-024-05532-4. BMC Plant Biol. 2024. PMID: 39198770 Free PMC article. - Epigenetic regulation of intragenic transposable elements impacts gene transcription in Arabidopsis thaliana.
Le TN, Miyazaki Y, Takuno S, Saze H. Le TN, et al. Nucleic Acids Res. 2015 Apr 30;43(8):3911-21. doi: 10.1093/nar/gkv258. Epub 2015 Mar 26. Nucleic Acids Res. 2015. PMID: 25813042 Free PMC article. - DNA methylation is critical for Arabidopsis embryogenesis and seed viability.
Xiao W, Custard KD, Brown RC, Lemmon BE, Harada JJ, Goldberg RB, Fischer RL. Xiao W, et al. Plant Cell. 2006 Apr;18(4):805-14. doi: 10.1105/tpc.105.038836. Epub 2006 Mar 10. Plant Cell. 2006. PMID: 16531498 Free PMC article. - The methylation cycle and its possible functions in barley endosperm development.
Radchuk VV, Sreenivasulu N, Radchuk RI, Wobus U, Weschke W. Radchuk VV, et al. Plant Mol Biol. 2005 Sep;59(2):289-307. doi: 10.1007/s11103-005-8881-1. Plant Mol Biol. 2005. PMID: 16247558 - Changes in gene expression in response to altered SHL transcript levels.
Müssig C, Altmann T. Müssig C, et al. Plant Mol Biol. 2003 Dec;53(6):805-20. doi: 10.1023/B:PLAN.0000023661.65248.4b. Plant Mol Biol. 2003. PMID: 15082927
References
- Bell CJ, Ecker JR. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;19:137–144. - PubMed
- Bender J, Fink GR. Epigenetic control of an endogenous gene family is revealed by a novel blue fluorescent mutant of Arabidopsis. Cell. 1995;83:725–734. - PubMed
- Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases