Selective induction of E2F1 in response to DNA damage, mediated by ATM-dependent phosphorylation - PubMed (original) (raw)
. 2001 Jul 15;15(14):1833-44.
Affiliations
- PMID: 11459832
- PMCID: PMC312742
Selective induction of E2F1 in response to DNA damage, mediated by ATM-dependent phosphorylation
W C Lin et al. Genes Dev. 2001.
Abstract
Previous work has established a role for p53 in triggering apoptosis in response to DNA damage; p53 also induces apoptosis in response to deregulation of the Rb cell cycle pathway. The latter event is consistent with a role for the Rb-regulated E2F1 protein as a specific inducer of apoptosis and p53 accumulation. We now show that DNA damage leads to a specific induction of E2F1 accumulation, dependent on ATM kinase activity and that the specificity of E2F1 induction reflects a specificity in the phosphorylation of E2F1 by ATM as well as the related kinase ATR. We identify a site for ATM/ATR phosphorylation in the amino terminus of E2F1 and we show that this site is required for ATM-mediated stabilization of E2F1. Finally, we also show that E2F1 is required for DNA damaged induced apoptosis in mouse thymocytes. We conclude that the cellular response to DNA damage makes use of signals from the Rb/E2F cell cycle pathway.
Figures
Figure 1
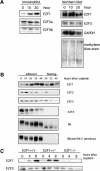
Induction of E2F1 accumulation following DNA damage. (A) Western blot analysis of E2F1 among a panel of human cell lines after 24 h treatment of adriamycin (adr), cisplatin (cisp), or etoposide (eto) shows induction of E2F1 protein regardless of their p53 or Rb status. The concentration of cisplatin was from 5 to 50 μM, adriamycin 50 to 500 nM, and etoposide 5 to 50 μM. H460, H358, H125, H520, and H596 are derived from lung cancer. Hep3B and HepG2 are hepatoma cell lines. HFF is a nontransformed human foreskin fibroblast cell line. H460 carries wild-type p53. H358 and Hep3B are _p53_-null. H125, H520, and H596 carry mutated p53. H596 and Hep3B have no Rb protein expression, whereas H460, H358, H125, and HepG2 carry normal Rb. Equal protein loading of each lane was confirmed by Ponceau-S staining. (B) Mouse embryo fibroblasts prepared from _p53_−/− embryos or wild-type littermates were treated with cisplatin (20 μM) for the indicated time. E2F1 in the lysates was detected by immunoblotting.
Figure 2
Specificity in posttranscriptional induction of E2F1 accumulation. (A) Measurements of E2F protein accumulation (immunoblot) and RNA accumulation (Northern blot) following treatment of H460 cells with cisplatin (50 μM). Two species of E2F3 protein are detected (E2F3a and E2F3b) that derive from alternate transcription initiation sites within the E2F3 locus (Leone et al. 2000). The two E2F3 transcripts are not resolved in this Northern analysis. (B) Western blot analysis of E2F and Rb proteins in the H460 human lung cancer cell line following treatment with cisplatin (50 μM). Rb protein is seen to be degraded to a truncated product that lacks the carboxyl terminus in the cells undergoing apoptosis, likely a result of caspase action (Tan and Wang 1998). (C) Western blot analysis of E2F in primary mouse embryonic fibroblasts (MEF). Primary MEF cultures were prepared from embryos with the indicated genotypes and then treated with cisplatin (20 μM).
Figure 3
Role of ATM in the induction of E2F1 following DNA damage. (A) Various human lymphoblastoid cell lines with the indicated ATM genotypes were treated with adriamycin (1 μM). E2F1 protein accumulation was assayed by Western blot analysis. For comparison, p53 protein accumulation was measured in the same samples. (B) Induction of E2F1 by etoposide. The lymphoblastoid cell lines were treated with etoposide (50 μM) for the indicated time and E2F1 was detected by immunoblotting.
Figure 4
Specific phosphorylation of E2F1 by ATM/ATR. (A) Wild-type ATM or catalytically inactive ATM (ATMki) was immunoprecipitated from transfected cells 293T cells and used to phosphorylate GST–p53 (aa 1–70), or GST–E2F1, GST–E2F2, and GST–E2F3 proteins in in vitro reactions. The upper panel depicts the 32P autoradiograph and the lower panel shows an immunoblot with GST antibody. (B) Similar assays as described in A were performed with wild-type ATR or catalytically inactive ATR (ATRki).
Figure 5
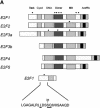
A basis for the specific phosphorylation of E2F1 by ATM and ATR. (A) Schematic depiction of the E2F family with potential sites for ATM/ATR phosphorylation. ATM/ATR consensus sites (S/T-Q) found within the E2F1–3 subfamily are indicated by the dots. The amino acid sequence containing the unique amino-terminal site in E2F1 is indicated at the bottom. (B) In vitro phosphorylation of E2F1 at serine 31. Wild-type E2F1 protein or the E2F1S31A mutant was prepared as described in Materials and Methods and used as a substrate for ATM/ATR kinase assays in vitro. The upper panel depicts the 32P autoradiograph after the kinase reaction whereas the lower panel depicts staining of the input proteins.
Figure 5
A basis for the specific phosphorylation of E2F1 by ATM and ATR. (A) Schematic depiction of the E2F family with potential sites for ATM/ATR phosphorylation. ATM/ATR consensus sites (S/T-Q) found within the E2F1–3 subfamily are indicated by the dots. The amino acid sequence containing the unique amino-terminal site in E2F1 is indicated at the bottom. (B) In vitro phosphorylation of E2F1 at serine 31. Wild-type E2F1 protein or the E2F1S31A mutant was prepared as described in Materials and Methods and used as a substrate for ATM/ATR kinase assays in vitro. The upper panel depicts the 32P autoradiograph after the kinase reaction whereas the lower panel depicts staining of the input proteins.
Figure 6
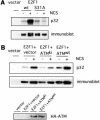
DNA damage induced phosphorylation of E2F1. (A) 293T cells were transfected with HA-E2F1 or HA-E2F1S31A and labeled with 32P orthophosphate in the absence or presence of neocarzinostatin (NCS) at 300 ng/ml. E2F1 protein was then immunoprecipitated with HA beads and phosphorylation was detected by autoradiography (top). Also shown is the protein level as seen by immunoblotting (bottom). (B) A role for ATM in DNA damage induced phosphorylation of E2F1. 293T cells were transfected with HA-E2F1 along with vector, HA-ATMwt, or HA-ATMki. The cells were then labeled with 32P orthophosphate in the absence or presence of NCS and E2F1 protein was immunoprecipitated and detected as in A. Expression of transfected HA-ATM was confirmed by immunoblotting the HA bead precipitate with ATM antibody (Ab-3).
Figure 7
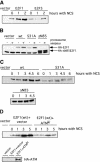
The ATM phosphorylation site in E2F1 is required for DNA damaged induction. (A) 293T cells were transfected with HA-E2F1 or HA-E2F3 and then treated with neocarzinostatin (NCS) for the indicated time. HA-tagged E2F proteins were detected by immunoblotting with an HA-specific antibody. (B) Degradation of E2F1 by the proteasome. 293T cells were transfected with HA-E2F1, HA-E2F1S31A, or HA-E2F1ΔN85 and then treated with MG-132 at 20 μM for 5 h. Proteins were detected by immunoblotting with an HA specific antibody. (C) 293T cells were transfected with HA-E2F1, HA-E2F1S31A, or HA-E2F1ΔN85 and then treated with NCS for the indicated time. Proteins were detected by immunoblotting with an HA specific antibody. (D) Induction of E2F1 in response to DNA damage requires ATM. 293T cells were transfected with HA-E2F1 along with HA-ATMKi or vector. The cells were then treated with NCS for the indicated time. Transfected E2F1 was detected with an HA-specific antibody (top). Expression of transfected ATMki protein was confirmed by immunoprecipitation with HA beads followed by immunoblotting with ATM antibody (Ab-3) (bottom).
Figure 8
A role for E2F1 in DNA damage-induced thymocyte death. (A) Western blot analysis of E2F1 protein induction in thymocytes analyzed 22 h following treatment with etoposide (2 μM). (B) Viability assays. Thymocytes were prepared from 4–8-week-old mice and cell viability was analyzed 20 h following treatment with etoposide at concentrations ranging from 2 to 500 μM. (C) Viability assays. Thymocytes were prepared from six month-old mice and cell viability was analyzed 20 h following treatment with etoposide at concentrations ranging from 2 to 200 μM. (D) Flow cytometry profiles following CD4 and CD8 immunostaining of thymocytes from E2F1+/+ or _E2F1_−/− mice at either eight wk of age or six mo of age.
Figure 9
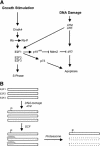
Role of E2F1 in the DNA damage response. (A) A link between the Rb/E2F pathway and the DNA damage response. In addition to the role of ATM and ATR in inducing p53 accumulation, the results presented here identify E2F1 as a target for ATM and ATR in response to DNA damage. The consequence of the E2F1 induction could be seen as a further augmentation of p53 accumulation through an activation of p19ARF. In addition, given the ability of E2F1 to induce the p73 gene, the action through E2F1 could lead to a p53-independent effect on apoptosis. (B) Potential mechanism for ATM/ATR-mediated stabilization of E2F1. Previous work has documented the role of an SCF complex in targeting an amino-terminal domain of E2F1, leading to ubiquitin-dependent proteasome degradation. The site for ATM/ATR phosphorylation lies within this domain; as such, we speculate that the phosphorylation might prevent the interaction of the SCF complex with E2F1 and thus prevent degradation.
Similar articles
- E2F1 uses the ATM signaling pathway to induce p53 and Chk2 phosphorylation and apoptosis.
Powers JT, Hong S, Mayhew CN, Rogers PM, Knudsen ES, Johnson DG. Powers JT, et al. Mol Cancer Res. 2004 Apr;2(4):203-14. Mol Cancer Res. 2004. PMID: 15140942 - ATM is a target for positive regulation by E2F-1.
Berkovich E, Ginsberg D. Berkovich E, et al. Oncogene. 2003 Jan 16;22(2):161-7. doi: 10.1038/sj.onc.1206144. Oncogene. 2003. PMID: 12527885 - Specificity in the activation and control of transcription factor E2F-dependent apoptosis.
Hallstrom TC, Nevins JR. Hallstrom TC, et al. Proc Natl Acad Sci U S A. 2003 Sep 16;100(19):10848-53. doi: 10.1073/pnas.1831408100. Epub 2003 Sep 3. Proc Natl Acad Sci U S A. 2003. PMID: 12954980 Free PMC article. - Life, death and E2F: linking proliferation control and DNA damage signaling via E2F1.
Rogoff HA, Kowalik TF. Rogoff HA, et al. Cell Cycle. 2004 Jul;3(7):845-6. doi: 10.4161/cc.3.7.976. Epub 2004 Jul 14. Cell Cycle. 2004. PMID: 15190206 Review. - The emerging role of E2F-1 in the DNA damage response and checkpoint control.
Stevens C, La Thangue NB. Stevens C, et al. DNA Repair (Amst). 2004 Aug-Sep;3(8-9):1071-9. doi: 10.1016/j.dnarep.2004.03.034. DNA Repair (Amst). 2004. PMID: 15279795 Review.
Cited by
- Jab1 is a specificity factor for E2F1-induced apoptosis.
Hallstrom TC, Nevins JR. Hallstrom TC, et al. Genes Dev. 2006 Mar 1;20(5):613-23. doi: 10.1101/gad.1345006. Epub 2006 Feb 15. Genes Dev. 2006. PMID: 16481464 Free PMC article. - Vanadate activated Akt and promoted S phase entry.
Zhang Z, Gao N, He H, Huang C, Luo J, Shi X. Zhang Z, et al. Mol Cell Biochem. 2004 Jan;255(1-2):227-37. doi: 10.1023/b:mcbi.0000007278.27936.8b. Mol Cell Biochem. 2004. PMID: 14971663 - Regulation of E2F1 by BRCT domain-containing protein TopBP1.
Liu K, Lin FT, Ruppert JM, Lin WC. Liu K, et al. Mol Cell Biol. 2003 May;23(9):3287-304. doi: 10.1128/MCB.23.9.3287-3304.2003. Mol Cell Biol. 2003. PMID: 12697828 Free PMC article. - Genes and pathways driving glioblastomas in humans and murine disease models.
Merlo A. Merlo A. Neurosurg Rev. 2003 Jul;26(3):145-58. doi: 10.1007/s10143-003-0267-8. Epub 2003 May 29. Neurosurg Rev. 2003. PMID: 12783270 Review. - E2F3 is a mediator of DNA damage-induced apoptosis.
Martinez LA, Goluszko E, Chen HZ, Leone G, Post S, Lozano G, Chen Z, Chauchereau A. Martinez LA, et al. Mol Cell Biol. 2010 Jan;30(2):524-36. doi: 10.1128/MCB.00938-09. Epub 2009 Nov 16. Mol Cell Biol. 2010. PMID: 19917728 Free PMC article.
References
- Banin S, Moyal L, Shieh S, Taya Y, Anderson CW, Chessa L, Smorodinsky NI, Prives C, Reiss Y, Shiloh Y, et al. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281:1674–1677. - PubMed
- Bates S, Phillips AC, Clark PA, Stott F, Peters G, Ludwig RL, Vousden KH. p14ARF links the tumour suppressors RB and p53. Nature. 1998;395:124–125. - PubMed
- Canman CE, Lim DS, Cimprich KA, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan MB, Siliciano JD. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–1679. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous