Bacteriophage T4 gene 41 helicase and gene 59 helicase-loading protein: a versatile couple with roles in replication and recombination - PubMed (original) (raw)
Review
Bacteriophage T4 gene 41 helicase and gene 59 helicase-loading protein: a versatile couple with roles in replication and recombination
C E Jones et al. Proc Natl Acad Sci U S A. 2001.
Abstract
Bacteriophage T4 uses two modes of replication initiation: origin-dependent replication early in infection and recombination-dependent replication at later times. The same relatively simple complex of T4 replication proteins is responsible for both modes of DNA synthesis. Thus the mechanism for loading the T4 41 helicase must be versatile enough to allow it to be loaded on R loops created by transcription at several origins, on D loops created by recombination, and on stalled replication forks. T4 59 helicase-loading protein is a small, basic, almost completely alpha-helical protein whose N-terminal domain has structural similarity to high mobility group family proteins. In this paper we review recent evidence that 59 protein recognizes specific structures rather than specific sequences. It binds and loads the helicase on replication forks and on three- and four-stranded (Holliday junction) recombination structures, without sequence specificity. We summarize our experiments showing that purified T4 enzymes catalyze complete unidirectional replication of a plasmid containing the T4 ori(uvsY) origin, with a preformed R loop at the position of the R loop identified at this origin in vivo. This replication depends on the 41 helicase and is strongly stimulated by 59 protein. Moreover, the helicase-loading protein helps to coordinate leading and lagging strand synthesis by blocking replication on the ori(uvsY) R loop plasmid until the helicase is loaded. The T4 enzymes also can replicate plasmids with R loops that do not have a T4 origin sequence, but only if the R loops are within an easily unwound DNA sequence.
Figures
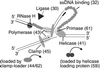
Figure 1
Model of a replication fork with bacteriophage T4 proteins. Numbers indicate the name of the gene encoding each protein. See text for description of the functions of the proteins.
Figure 2
Ribbon diagrams of the crystal structure of the T4 gene 59 helicase-loading protein showing its structural similarity with HMG proteins. (A) and (B) are, respectively, “top” and “side” view rainbow-colored ribbon diagrams of 59 protein (PDB 1C1K, ref. 8) from the blue N terminus to the red C terminus. S1–2 are β-sheets and H1–13 are α-helices. The line between H6 and H7 indicates the junction between the N and C domains of the protein. (C) Superposition of the H1, H2, and H3 α-helices of T4 59 helicase-loading protein (blue) and rat HMG1 protein (green, PDB 1AAB, ref. 20) and with the DNA (red) from the model of lymphoid enhancer-binding factor LEF1-DNA, (PDB 2LEF, ref. 21). Adapted from Mueser et al. (8).
Figure 3
Fork DNA structures bound by T4 59 helicase-loading protein and unwound by the 41 helicase loaded by 59 protein. (A) A speculative model of T4 gene 59 helicase-loading protein bound on a DNA replication fork. This model is taken from ref. and was adapted from Mueser et al. (8). It is based on the distribution of hydrophobic and basic residues on the surface of 59 protein and on the assumption that the HMG-like region of its N domain binds and unstacks the duplex (red and green) ahead of the fork, as shown for HMG proteins (17, 19). The leading strand duplex [template (green)/primer (blue)] is docked on the bottom surface of the C domain. A long segment of single-stranded DNA, representing the lagging strand (red), traverses the shallow groove between the N and C domains. A helicase monomer (light blue oval) is proposed to bind to 59 protein between the lagging and leading strand arms (8). The other subunits of the hexameric helicase (not shown) would surround the lagging strand. (B) 59 protein binds to forks with single or duplex arms, but cannot load the helicase without a single-stranded gap of more than 5 nt on the lagging strand template. The fork DNA structures are drawn in the same orientation as the fork DNA on the model of 59 protein in A. The hexameric helicase (circle) is placed on the strand that would occupy the position of the fork lagging strand, if each of these DNAs binds to 59 protein as predicted by the model in A. Arrowheads represent the 3′ end of each strand; * marks the position of 32P label in the substrates tested. [Reproduced with permission from ref. (Copyright 2000, American Society for Biochemistry and Molecular Biology).]
Figure 4
T4 59 protein stimulates unwinding of cruciform DNA substrates by 41 DNA helicase. (A) Diagram showing the expected unwinding products and positions of the helicase (circle) on a four-stranded cruciform and three-stranded invasion structure, assuming that 59 protein binds to the fork portions of these structures and loads the helicase as predicted by the speculative model in Fig. 3_A_. Arrowheads represent the 3′ end of each strand; * marks the position of 32P label in the substrates tested. Strand exchange was not possible because the arms of the structures were not homologous. (B) Unwinding of cruciform and strand invasion structures. Helicase was present at a final concentration of 200 nM (monomer) where indicated. DNA was present at 10 nM. Adapted from ref. .
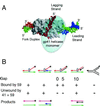
Figure 5
Replication of a plasmid with a preformed R loop at the T4 ori(uvsY) replication origin by T4 replication proteins in vitro. (A) Diagram of the pKK405 plasmid (57), which contains a 1.35-kb fragment of T4 DNA (solid line) with the ori(uvsY) replication origin in a pBR322-based vector (dashed line). As indicated in the text, this origin contains the PuvsY transcription promoter adjoining a DUE. The R loop in the plasmid was formed by annealing a 104-b RNA corresponding to −5 to +99 relative to the transcription start site (10). (B) T4 replication proteins required for unidirectional replication from the pKK405 R loop plasmid, giving nicked circular products. [B, Reproduced with permission from ref. (Copyright 2001, Elsevier Science).]
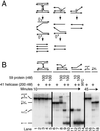
Figure 6
Replication of the R loop plasmid with the T4 ori(uvsY) origin requires 41 helicase and is strongly stimulated by the 59 helicase-loading protein. (A) Model showing the roles of the 59 helicase-loading protein, 41 helicase, and 61 primase in unidirectional replication from T4 ori(uvsY). 59 Protein binds ahead of the polymerase holoenzyme elongating the RNA in the R loop and loads the helicase essential for rapid leading strand synthesis. 59 Protein inhibits leading strand synthesis when there is no helicase. (B) Neutral agarose gel of the products of unidirectional continuous replication of the pKK405 ori(uvsY) R loop plasmid. The complete reactions contained the T4 replication proteins listed in Fig. 5_B_. 59 Helicase-loading protein, 41 helicase, and 61 primase were omitted as indicated. (C) Alkaline agarose gel of the replication of the pKK405 R loop plasmid in a two-stage reaction in which only the leading strand is labeled. The RNA was 3′ end-labeled by limited synthesis with dATP, dTTP, 32P-dCTP in a first-stage reaction that required T4 polymerase, clamp, clamp loader, and 32 protein. 41 Helicase, 59 helicase loader, 61 primase, and topoisomerase then were added along with dGTP and enough unlabeled dCTP to prevent further 32P-dCTP incorporation. The indicated enzymes were omitted in the second stage. T4 topoisomerase was present in all reactions. See text for description of the inhibition by 59 helicase-loading protein when 41 helicase is omitted and of the aberrant products made without the 61 primase. [B and C, Reproduced with permission from ref. (Copyright 2001, Elsevier Science).]
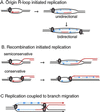
Figure 7
Models for the role of 41 helicase in origin-initiated and recombination-initiated replication. Helicase hexamers (circles) are shown on the strands they surround. Arrows above the circles show the direction of helicase movement. Arrowheads indicate the 3′ strand ends. RNA and invading DNA molecules are in red. Newly synthesized DNA is a dashed line in blue. (A) One or two helicases are required, respectively, for unidirectional or bidirectional replication from an origin. The helicase at each fork opens the duplex ahead of the polymerase and enables primer synthesis by the primase. (B) Semiconservative and conservative replication at forks initiated by recombination. In semiconservative replication, the displaced strand serves as the lagging strand template, and the helicase role is like that at an origin. In conservative replication, the invading strand serves as the lagging strand template. The helicase at the right catalyzes the branch migration needed for reannealing the duplex behind the leading strand and also interacts with the primase on the invading strand. (C) Semiconservative replication coupled to branch migration at a four-way junction. Helicase at the fork opens the duplex and interacts with primase. Branch migration is catalyzed by 5′ to 3′ movement of helicase on the four-way junction behind the leading strand polymerase. Two helicase hexamers are shown on the junction because the products of unwinding a cruciform with nonhomologous arms are most easily explained by two 41 helicases unwinding the cruciform simultaneously (see Fig. 4). However, it remains to be determined whether a single hexamer is sufficient for strand exchange at a Holliday junction. See text for further discussion. Adapted from ref. .
Similar articles
- Bacteriophage T4 proteins replicate plasmids with a preformed R loop at the T4 ori(uvsY) replication origin in vitro.
Nossal NG, Dudas KC, Kreuzer KN. Nossal NG, et al. Mol Cell. 2001 Jan;7(1):31-41. doi: 10.1016/s1097-2765(01)00152-6. Mol Cell. 2001. PMID: 11172709 - Control of helicase loading in the coupled DNA replication and recombination systems of bacteriophage T4.
Branagan AM, Klein JA, Jordan CS, Morrical SW. Branagan AM, et al. J Biol Chem. 2014 Jan 31;289(5):3040-54. doi: 10.1074/jbc.M113.505842. Epub 2013 Dec 14. J Biol Chem. 2014. PMID: 24338568 Free PMC article. - Mediator proteins orchestrate enzyme-ssDNA assembly during T4 recombination-dependent DNA replication and repair.
Bleuit JS, Xu H, Ma Y, Wang T, Liu J, Morrical SW. Bleuit JS, et al. Proc Natl Acad Sci U S A. 2001 Jul 17;98(15):8298-305. doi: 10.1073/pnas.131007498. Proc Natl Acad Sci U S A. 2001. PMID: 11459967 Free PMC article. Review. - Initiation of bacteriophage T4 DNA replication and replication fork dynamics: a review in the Virology Journal series on bacteriophage T4 and its relatives.
Kreuzer KN, Brister JR. Kreuzer KN, et al. Virol J. 2010 Dec 3;7:358. doi: 10.1186/1743-422X-7-358. Virol J. 2010. PMID: 21129203 Free PMC article. Review.
Cited by
- Phage single-stranded DNA-binding protein or host DNA damage triggers the activation of the AbpAB phage defense system.
Sasaki T, Takita S, Fujishiro T, Shintani Y, Nojiri S, Yasui R, Yonesaki T, Otsuka Y. Sasaki T, et al. mSphere. 2023 Dec 20;8(6):e0037223. doi: 10.1128/msphere.00372-23. Epub 2023 Oct 26. mSphere. 2023. PMID: 37882551 Free PMC article. - Bi- and Multi-directional Gene Transfer in the Natural Populations of Polyvalent Bacteriophages, and Their Host Species Spectrum Representing Foodborne Versus Other Human and/or Animal Pathogens.
Gabashvili E, Kobakhidze S, Koulouris S, Robinson T, Kotetishvili M. Gabashvili E, et al. Food Environ Virol. 2021 Jun;13(2):179-202. doi: 10.1007/s12560-021-09460-6. Epub 2021 Jan 23. Food Environ Virol. 2021. PMID: 33484405 - Characterization and comparative genomic analysis of virulent and temperate Bacillus megaterium bacteriophages.
Sharaf A, Oborník M, Hammad A, El-Afifi S, Marei E. Sharaf A, et al. PeerJ. 2018 Dec 10;6:e5687. doi: 10.7717/peerj.5687. eCollection 2018. PeerJ. 2018. PMID: 30581654 Free PMC article. - In vitro reconstitution of DNA replication initiated by genetic recombination: a T4 bacteriophage model for a type of DNA synthesis important for all cells.
Barry J, Wong ML, Alberts B. Barry J, et al. Mol Biol Cell. 2019 Jan 1;30(1):146-159. doi: 10.1091/mbc.E18-06-0386. Epub 2018 Nov 7. Mol Biol Cell. 2019. PMID: 30403545 Free PMC article. - Bacteriophage T5 gene D10 encodes a branch-migration protein.
Wong IN, Sayers JR, Sanders CM. Wong IN, et al. Sci Rep. 2016 Dec 23;6:39414. doi: 10.1038/srep39414. Sci Rep. 2016. PMID: 28009009 Free PMC article.
References
- Kreuzer K N, Morrical S W. In: Molecular Biology of Bacteriophage T4. Karem J, editor. Washington, DC: Am. Soc. Microbiol.; 1994. pp. 28–42.
- Kreuzer K N. Trends Biochem Sci. 2000;25:165–173. - PubMed
- Mosig G, Colowick N, Gruidl M E, Chang A, Harvey A J. FEMS Microbiol Rev. 1995;17:83–98. - PubMed
- Mosig G. Annu Rev Genet. 1998;32:379–413. - PubMed
- Nossal N G. In: Molecular Biology of Bacteriophage T4. Karem J, editor. Washington, DC: Am. Soc. Microbiol.; 1994. pp. 43–53.
Publication types
MeSH terms
Substances
Grants and funding
- R37 GM034622/GM/NIGMS NIH HHS/United States
- F32 GM19000/GM/NIGMS NIH HHS/United States
- R01 GM034622/GM/NIGMS NIH HHS/United States
- GM34622/GM/NIGMS NIH HHS/United States
- F32 GM019000/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources