The elongation domain of ELL is dispensable but its ELL-associated factor 1 interaction domain is essential for MLL-ELL-induced leukemogenesis - PubMed (original) (raw)
The elongation domain of ELL is dispensable but its ELL-associated factor 1 interaction domain is essential for MLL-ELL-induced leukemogenesis
R T Luo et al. Mol Cell Biol. 2001 Aug.
Abstract
The MLL-ELL chimeric gene is the product of the (11;19)(q23p13.1) translocation associated with de novo and therapy-related acute myeloid leukemias (AML). ELL is an RNA polymerase II elongation factor that interacts with the recently identified EAF1 (ELL associated factor 1) protein. EAF1 contains a limited region of homology with the transcriptional activation domains of three other genes fused to MLL in leukemias, AF4, LAF4, and AF5q31. Using an in vitro transformation assay of retrovirally transduced myeloid progenitors, we conducted a structure-function analysis of MLL-ELL. Whereas the elongation domain of ELL was dispensable, the EAF1 interaction domain of ELL was critical to the immortalizing properties of MLL-ELL in vitro. To confirm these results in vivo, we transplanted mice with bone marrow transduced with MLL fused to the minimal EAF1 interaction domain of ELL. These mice all developed AML, with a longer latency than mice transplanted with the wild-type MLL-ELL fusion. Based on these results, we generated a heterologous MLL-EAF1 fusion gene and analyzed its transforming potential. Strikingly, we found that MLL-EAF1 immortalized myeloid progenitors in the same manner as that of MLL-ELL. Furthermore, transplantation of bone marrow transduced with MLL-EAF1 induced AML with a shorter latency than mice transplanted with the MLL-ELL fusion. Taken together, these results indicate that the leukemic activity of MLL-ELL requires the EAF1 interaction domain of ELL, suggesting that the recruitment by MLL of a transactivation domain similar to that in EAF1 or the AF4/LAF4/AF5q31 family may be a critical common feature of multiple 11q23 translocations. In addition, these studies support a critical role for MLL partner genes and their protein-protein interactions in 11q23 leukemogenesis.
Figures
FIG. 1
Determination of the sequences of ELL essential to the immortalization of hematopoietic progenitor cells by MLL-ELL in a myeloid clonogenic assay. The bars in the right column represent the number of tertiary colonies generated by the respective mutants depicted in the left column.
FIG. 2
Western blot and RT-PCR analysis of the expression of the various constructs. The upper panels show expression of the various MLL fusion protein constructs in transiently transfected BOSC cells using antibodies to MLL, ELL, or EAF1. The lower panels show RT-PCR expression analysis in the transduced myeloid clonogenic cells harvested from primary methylcellulose cultures. Amplification from reverse transcribed cDNA is indicated by a plus symbol (+). To exclude amplification of integrated retroviral genomic DNA, a no-RT control is indicated by a minus symbol (−).
FIG. 3
Mapping of the EAF1 interaction domain within ELL. (A) The human 293 cell line was transfected with FLAG-tagged constructs containing different regions of ELL. Western blot analysis of cell lysates confirmed the expression of each of the constructs. (B) Immunoprecipitation of cell lysates was performed with the FLAG antibody, followed by stringent washes of the immunoprecipitated complexes. The minimal domain that coprecipitated with endogenous EAF1 mapped to amino acids 508 to 621 within ELL. Endogenous EAF1 migrates at approximately 43 kDa.
FIG. 4
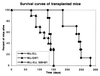
Survival curves of the mice transplanted with BM progenitors transduced by the MLL-ELL, MLL-ELL508–621, and MLL-EAF1 encoding vectors.
FIG. 5
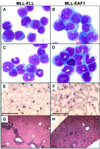
Morphology of the neoplastic cells in MLL-ELL (left column) and MLL-EAF1 leukemic mice (right column). Wright-Giemsa staining of BM cytospin preparation (A, B, C, and D). (A) MLL-ELL mouse with acute monoblastic leukemia (poorly differentiated). (B) MLL-EAF1 mouse with acute monocytic leukemia (differentiated). (C and D) MLL-ELL and MLL-EAF1 mice, respectively, with acute myelomonocytic leukemia. (E and F) PB smears showing circulating blast cells. (G and H) Histological analysis of the liver showing infiltration by leukemia cells. Bar, 300 μm.
FIG. 6
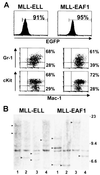
(A) Immunophenotype of the leukemic BM of MLL-ELL (left) and MLL-EAF1 (right) mice. The histograms show the expression of the EGFP marker, and the region used to gate the EGFP-positive population is indicated, along with the percentage of cells it comprises. The dot plots represent the Mac-1 versus Gr-1 or cKit staining of the EGFP-expressing cells. Percentage values correspond to the content of the adjacent quadrants. (B) Southern blot analysis of spleen DNA obtained from leukemic mice, digested with _Bam_HI, and probed with an MLL cDNA fragment. As indicated by the arrows, one to three integration sites could be detected, indicating that the leukemias were mono- or pauciclonal in the MLL-ELL and the MLL-EAF1 mice. A 9-kb band is also detected, corresponding to the endogenous murine Mll gene.
FIG. 7
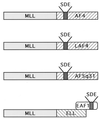
Model of 11q23 leukemogenesis involving AF4 family members and ELL. A subset of 11q23 translocations involves a fusion of MLL to the AF4, LAF4 and AF5q31 genes that contain transcriptional activation domain rich in serine (S), aspartic acid (D), and glutamic acid (E) residues. In the MLL-ELL fusion that results from the t(11;19)(q23;p13.1), ELL retains its interaction domain with EAF1, a transcriptional activator with homology to the serine (S)-, aspartic acid (D)-, and glutamic acid (E)-rich activation domains of AF4, LAF4, and AF5q31.
FIG. 8
CLUSTALW alignment of EAF1 with LAF4, AF5q31, and AF4. Amino acid identity is indicated by dark gray boxes, and amino acid similarity is indicated by light gray boxes.
Similar articles
- EAF1, a novel ELL-associated factor that is delocalized by expression of the MLL-ELL fusion protein.
Simone F, Polak PE, Kaberlein JJ, Luo RT, Levitan DA, Thirman MJ. Simone F, et al. Blood. 2001 Jul 1;98(1):201-9. doi: 10.1182/blood.v98.1.201. Blood. 2001. PMID: 11418481 - ELL-associated factor 2 (EAF2), a functional homolog of EAF1 with alternative ELL binding properties.
Simone F, Luo RT, Polak PE, Kaberlein JJ, Thirman MJ. Simone F, et al. Blood. 2003 Mar 15;101(6):2355-62. doi: 10.1182/blood-2002-06-1664. Epub 2002 Nov 21. Blood. 2003. PMID: 12446457 - A carboxy-terminal domain of ELL is required and sufficient for immortalization of myeloid progenitors by MLL-ELL.
DiMartino JF, Miller T, Ayton PM, Landewe T, Hess JL, Cleary ML, Shilatifard A. DiMartino JF, et al. Blood. 2000 Dec 1;96(12):3887-93. Blood. 2000. PMID: 11090074 - Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins.
Ayton PM, Cleary ML. Ayton PM, et al. Oncogene. 2001 Sep 10;20(40):5695-707. doi: 10.1038/sj.onc.1204639. Oncogene. 2001. PMID: 11607819 Review. - The biological and clinical significance of MLL abnormalities in haematological malignancies.
Mitterbauer-Hohendanner G, Mannhalter C. Mitterbauer-Hohendanner G, et al. Eur J Clin Invest. 2004 Aug;34 Suppl 2:12-24. doi: 10.1111/j.0960-135X.2004.01366.x. Eur J Clin Invest. 2004. PMID: 15291802 Review.
Cited by
- The little elongation complex regulates small nuclear RNA transcription.
Smith ER, Lin C, Garrett AS, Thornton J, Mohaghegh N, Hu D, Jackson J, Saraf A, Swanson SK, Seidel C, Florens L, Washburn MP, Eissenberg JC, Shilatifard A. Smith ER, et al. Mol Cell. 2011 Dec 23;44(6):954-65. doi: 10.1016/j.molcel.2011.12.008. Mol Cell. 2011. PMID: 22195968 Free PMC article. - ELL and EAF1 are Cajal body components that are disrupted in MLL-ELL leukemia.
Polak PE, Simone F, Kaberlein JJ, Luo RT, Thirman MJ. Polak PE, et al. Mol Biol Cell. 2003 Apr;14(4):1517-28. doi: 10.1091/mbc.e02-07-0394. Mol Biol Cell. 2003. PMID: 12686606 Free PMC article. - RNA Polymerase II-Dependent Transcription Initiated by Selectivity Factor 1: A Central Mechanism Used by MLL Fusion Proteins in Leukemic Transformation.
Yokoyama A. Yokoyama A. Front Genet. 2019 Jan 14;9:722. doi: 10.3389/fgene.2018.00722. eCollection 2018. Front Genet. 2019. PMID: 30693017 Free PMC article. Review. - Zebrafish eaf1 and eaf2/u19 mediate effective convergence and extension movements through the maintenance of wnt11 and wnt5 expression.
Liu JX, Hu B, Wang Y, Gui JF, Xiao W. Liu JX, et al. J Biol Chem. 2009 Jun 12;284(24):16679-16692. doi: 10.1074/jbc.M109.009654. Epub 2009 Apr 20. J Biol Chem. 2009. PMID: 19380582 Free PMC article. - Pirin down-regulates the EAF2/U19 protein and alleviates its growth inhibition in prostate cancer cells.
Qiao Z, Wang D, Hahn J, Ai J, Wang Z. Qiao Z, et al. Prostate. 2014 Feb;74(2):113-20. doi: 10.1002/pros.22729. Epub 2013 Nov 23. Prostate. 2014. PMID: 24272884 Free PMC article.
References
- Aso T, Lane W S, Conaway J W, Conaway R C. Elongin (SIII): a multisubunit regulator of elongation by RNA polymerase II. Science. 1995;269:1439–1443. - PubMed
- Butler L H, Slany R, Cui X, Cleary M L, Mason D Y. The HRX proto-oncogene product is widely expressed in human tissues and localizes to nuclear structures. Blood. 1997;89:3361–3370. - PubMed
- Bernard O A, Berger R. Molecular basis of 11q23 rearrangements in hematopoietic malignant proliferations. Genes Chromosomes Cancer. 1995;13:75–85. - PubMed
- Corral J, Lavenir I, Impey H, Warren A J, Forster A, Larson T A, Bell S, McKenzie A N, King G, Rabbitts T H. An Mll-Af9 fusion gene made by homologous recombination causes acute leukemia in chimeric mice: a method to create fusion oncogenes. Cell. 1996;85:853–861. - PubMed
- DiMartino J F, Cleary M L. MLL rearrangements in haematological malignancies: lessons from clinical and biological studies. Br J Haematol. 1999;106:614–626. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases