Arg3.1/Arc mRNA induction by Ca2+ and cAMP requires protein kinase A and mitogen-activated protein kinase/extracellular regulated kinase activation - PubMed (original) (raw)
Arg3.1/Arc mRNA induction by Ca2+ and cAMP requires protein kinase A and mitogen-activated protein kinase/extracellular regulated kinase activation
R Waltereit et al. J Neurosci. 2001.
Abstract
Long-term potentiation (LTP) is a cellular model for persistent synaptic plasticity in the mammalian brain. Like several forms of memory, long-lasting LTP requires cAMP-mediated activation of protein kinase A (PKA) and is dependent on gene transcription. Consequently, activity-dependent genes such as c-fos that contain cAMP response elements (CREs) in their 5' regulatory region have been studied intensely. More recently, arg3.1/arc became of interest, because after synaptic stimulation, arg3.1/arc mRNA is rapidly induced and distributed to dendritic processes and may be locally translated there to facilitate synapse-specific modifications. However, to date nothing is known about the signaling mechanisms involved in the induction of this gene. Here we report that arg3.1/arc is robustly induced with LTP stimulation even at intensities that are not sufficient to activate c-fos expression. Unlike c-fos, the 5' regulatory region of arg3.1/arc does not contain a CRE consensus sequence and arg3.1/arc is unresponsive to cAMP in NIH3T3 and Neuro2a cells. However, in PC12 cells and primary cultures of hippocampal neurons, arg3.1/arc can be induced by cAMP and calcium. This induction requires the activity of PKA and mitogen-activated protein kinase, suggesting a neuron-specific pathway for the activation of arg3.1/arc expression.
Figures
Fig. 1.
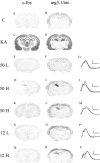
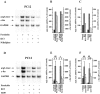
Comparison of arg3.1/arc and c-_fos_mRNA levels after kainic acid-induced seizures and LTP-producing stimulation. Coronal sections were assayed for arg3.1/arc and c-fos mRNA using in situ hybridization with antisense RNA probes. Representative autoradiographs of three independent experiments are shown. A, B, One hour after saline injection. C, D, One hour after kainic acid (10 mg/kg)-induced seizures.E, F, One hour after 50-train LFS of the perforant path in freely moving rats. H,I, K, L, One hour after 50-train HFS (n = 5). N, O, One hour after 12-train LFS. Q, R, One hour after 12-train HFS. G, J,M, P, S, Superimposed field potentials before and 1 hr after LFS or HFS, respectively. Calibration: 5 mV, 5 msec.
Fig. 2.
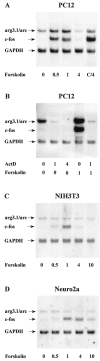
Depolarization regulates arg3.1/arc and c-fos mRNA levels in PC12 cells. Autoradiograph of Northern blot analysis of arg3.1/arc and c-_fos_transcripts. Four independent experiments were conducted (n = 4). Six micrograms of RNA isolated from PC12 cells were loaded per lane. The blot was hybridized to a probe specific for arg3.1/arc and a probe specific for c-fos. Hybridization to a probe specific for GAPDH was used as a loading control. Numbers below the lanes indicate the period of 60 m
m
KCl exposure in hours. Lane C/4, RNA isolated 4 hr after exposure to KCl in the presence of cycloheximide (CHX, 10 μg/ml). Lane C, RNA isolated 4 hr after exposure to CHX only. Note that the increase in arg3.1/arc mRNA levels is maintained for 4 hr and not superinduced by CHX. In contrast, c-fos mRNA is only transiently induced but strongly superinduced in the presence of CHX.
Fig. 3.
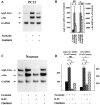
Regulation of arg3.1/arc and c-_fos_transcription by cAMP. Autoradiographs are of Northern blots. RNA amounts and labels are as in Figure 2. Cells were stimulated with the adenylyl cyclase activator forskolin (50 μ
m
).A, B, RNA isolated from PC12 cells (n = 4). Where indicated, cells in _B_were exposed to 3 m
m
of the transcription inhibitor actinomycin D (ActD), 1.25 and 4.25 hr before lysis, or 15 min before stimulation with forskolin, respectively.C, RNA isolated from NIH3T3 fibroblasts (n = 2). D, RNA isolated from Neuro2a neuroblastoma cells (n = 2). Note that arg3.1/arc is induced by forskolin in PC12 cells but not in NIH3T3 or Neuro2a cells.
Fig. 4.
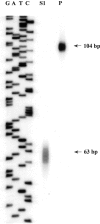
S1 nuclease mapping of the transcription start site. Right lane (P), Undigested S1-arg3.1/arc probe. Middle lane (S1), S1 nuclease-digested S1-arg3.1/arc oligonucleotide after hybridization with total RNA prepared from 60 min serum-stimulated NIH3T3 cells. The sequence of M13 on the left served as a length marker.
Fig. 5.
Sequence of the arg3.1/arc promoter region. The transcription start site is marked with asterisks. Promoter element consensus sequences are boxed. For a detailed description of the regulatory elements, refer to Results. A satellite sequence is underlined. The first codons of the reading frame are translated.
Fig. 6.
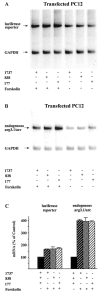
Response of the arg3.1/arc promoter to forskolin. Arg3.1/arc promoter deletions extending from position −1737 to +250, −834 to +250, and −177 to +250 were fused to a luciferase reporter. The reporter constructs were used to stably transfect PC12 cells. Cells were stimulated with forskolin for 1 hr. Inducibility of the arg3.1/arc promoter deletion constructs and that of the endogenous arg3.1/arc promoter was determined by Northern blot analyses.A, Autoradiograph of a Northern blot hybridized with a probe specific for luciferase to measure the inducibility of the transfected arg3.1/arc promoter deletions. Hybridization with a probe specific for GAPDH was used as a loading control.Numbers on the left indicate the 5′ deletion endpoints of the arg3.1/arc promoter constructs. Plus signs in the corresponding line indicate the presence of the respective constructs. The first three lanes from the_left_ contained RNA from forskolin-stimulated cells; the_three lanes_ on the right contained RNA from mock-stimulated cells. B, Autoradiograph of a Northern blot hybridized with a probe specific for arg3.1/arc to measure the inducibility of the endogenous arg3.1/arc promoter and a probe specific for GAPDH. Labels are as in A.C, Quantification of Northern blots (n = 3). Hybridization signals were normalized against mock-stimulated control and GAPDH signals. Error bars represent the SEM; p < 0.05 compared with control (two-tailed Student's t test). Note that transient transfection experiments indicated that the observed weak residual inducibility represents a function of the basic vector itself and not of the inserted arg3.1/arc sequences (data not shown).
Fig. 7.
Induced levels of arg3.1/arc and c-fos mRNA can be blocked by calcium channel and protein kinase A inhibitors in PC12 cells. PC12 cells were pretreated for 30 min with the indicated inhibitors and subsequently stimulated by the addition of either KCl or forskolin. A,D, Autoradiographs of Northern blots hybridized to probes specific for arg3.1/arc, c-fos, and GAPDH.B, E, Quantification of hybridization signals after forskolin stimulation. C,F, Quantification of hybridization signals after KCl depolarization. A_–_C, Effect of the calcium channel blocker nifedipine (10 μ
m
) on induced arg3.1/arc and c-fos mRNA levels (_n_= 2). D_–_F, Effect of the protein kinase A blocker H-89 (20 μ
m
) on induced arg3.1/arc and c-fos mRNA levels (n = 3). Error bars represent SEM; *p < 0.05 (two-tailed Student's _t_ test); _NS_, not significant,_p_ > 0.05. The induction of arg3.1/arc mRNA by forskolin and KCl was completely blocked by the PKA inhibitor.
Fig. 8.
Forskolin-induced arg3.1/arc mRNA levels are blocked by MAPK/ERK kinase inhibitor in PC12 cells and primary cultures of hippocampal neurons. PC12 cells and primary cultures of hippocampal neurons were pretreated with the MAPK/ERK kinase blocker PD098059 (50 μ
m
) and subsequently stimulated by forskolin (n = 3). Hippocampal neurons were also pretreated with the PKA blocker H-89 (compare with Fig.7_D_,E). A,C, Autoradiographs of Northern blots loaded with RNA from PC12 cells (A) and RNA from hippocampal neurons (C). Hybridization was with probes specific for arg3.1/arc, c-fos, and GAPDH.B,D, Quantification of arg3.1/arc and c-fos hybridization signals from analyses of PC12 cells and hippocampal neurons, respectively. Error bars represent SEM; *p < 0.05 (two-tailed Student's_t_ test); _NS_, not significant,_p_ > 0.05. Although the induction of arg3.1/arc mRNA by forskolin was blocked in PC12 cells and hippocampal neurons by the MAPK/ERK kinase inhibitor, induction of c-fos mRNA was affected to a much lesser extent.
Similar articles
- Actin polymerization and ERK phosphorylation are required for Arc/Arg3.1 mRNA targeting to activated synaptic sites on dendrites.
Huang F, Chotiner JK, Steward O. Huang F, et al. J Neurosci. 2007 Aug 22;27(34):9054-67. doi: 10.1523/JNEUROSCI.2410-07.2007. J Neurosci. 2007. PMID: 17715342 Free PMC article. - Brain-derived neurotrophic factor induces long-term potentiation in intact adult hippocampus: requirement for ERK activation coupled to CREB and upregulation of Arc synthesis.
Ying SW, Futter M, Rosenblum K, Webber MJ, Hunt SP, Bliss TV, Bramham CR. Ying SW, et al. J Neurosci. 2002 Mar 1;22(5):1532-40. doi: 10.1523/JNEUROSCI.22-05-01532.2002. J Neurosci. 2002. PMID: 11880483 Free PMC article. - Arc/Arg3.1 translation is controlled by convergent N-methyl-D-aspartate and Gs-coupled receptor signaling pathways.
Bloomer WAC, VanDongen HMA, VanDongen AMJ. Bloomer WAC, et al. J Biol Chem. 2008 Jan 4;283(1):582-592. doi: 10.1074/jbc.M702451200. Epub 2007 Nov 2. J Biol Chem. 2008. PMID: 17981809 - Inverse synaptic tagging: An inactive synapse-specific mechanism to capture activity-induced Arc/arg3.1 and to locally regulate spatial distribution of synaptic weights.
Okuno H, Minatohara K, Bito H. Okuno H, et al. Semin Cell Dev Biol. 2018 May;77:43-50. doi: 10.1016/j.semcdb.2017.09.025. Epub 2017 Sep 23. Semin Cell Dev Biol. 2018. PMID: 28939038 Review. - Molecular mechanism of neuronal plasticity: induction and maintenance of long-term potentiation in the hippocampus.
Miyamoto E. Miyamoto E. J Pharmacol Sci. 2006;100(5):433-42. doi: 10.1254/jphs.cpj06007x. J Pharmacol Sci. 2006. PMID: 16799259 Review.
Cited by
- Nicotinic neuromodulation in auditory cortex requires MAPK activation in thalamocortical and intracortical circuits.
Intskirveli I, Metherate R. Intskirveli I, et al. J Neurophysiol. 2012 May;107(10):2782-93. doi: 10.1152/jn.01129.2011. Epub 2012 Feb 22. J Neurophysiol. 2012. PMID: 22357798 Free PMC article. - A model of the roles of essential kinases in the induction and expression of late long-term potentiation.
Smolen P, Baxter DA, Byrne JH. Smolen P, et al. Biophys J. 2006 Apr 15;90(8):2760-75. doi: 10.1529/biophysj.105.072470. Epub 2006 Jan 13. Biophys J. 2006. PMID: 16415049 Free PMC article. - A form of perforant path LTP can occur without ERK1/2 phosphorylation or immediate early gene induction.
Steward O, Huang F, Guzowski JF. Steward O, et al. Learn Mem. 2007 Jun 11;14(6):433-45. doi: 10.1101/lm.554607. Print 2007 Jun. Learn Mem. 2007. PMID: 17562895 Free PMC article. - Experience-dependent coincident expression of the effector immediate-early genes arc and Homer 1a in hippocampal and neocortical neuronal networks.
Vazdarjanova A, McNaughton BL, Barnes CA, Worley PF, Guzowski JF. Vazdarjanova A, et al. J Neurosci. 2002 Dec 1;22(23):10067-71. doi: 10.1523/JNEUROSCI.22-23-10067.2002. J Neurosci. 2002. PMID: 12451105 Free PMC article. - Converging signal on ERK1/2 activity regulates group I mGluR-mediated Arc transcription.
Wang Y, Zheng F, Zhou X, Sun Z, Wang H. Wang Y, et al. Neurosci Lett. 2009 Aug 21;460(1):36-40. doi: 10.1016/j.neulet.2009.05.023. Epub 2009 May 14. Neurosci Lett. 2009. PMID: 19446601 Free PMC article.
References
- Abel T, Nguyen PV, Barad M, Deuel TA, Kandel ER, Bourtchouladze R. Genetic demonstration of a role for PKA in the late phase of LTP and in hippocampus-based long-term memory. Cell. 1997;88:615–626. - PubMed
- Abraham WC, Mason SE, Demmer J, Williams JM, Richardson CL, Tate WP, Lawlor PA, Dragunow M. Correlations between immediate early gene induction and the persistence of long-term potentiation. Neuroscience. 1993;56:717–727. - PubMed
- Alessi DR, Cuenda A, Cohen P, Dudley DT, Saltiel AR. PD 098059 is a specific inhibitor of the activation of mitogen-activated protein kinase kinase in vitro and in vivo. J Biol Chem. 1995;270:27489–27494. - PubMed
- Altschuler D, Lapetina EG. Mutational analysis of the cAMP-dependent protein kinase-mediated phosphorylation site of Rap1b. J Biol Chem. 1993;268:7527–7531. - PubMed
- Atkins CM, Selcher JC, Petraitis JJ, Trzaskos JM, Sweatt JD. The MAPK cascade is required for mammalian associative learning. Nat Neurosci. 1998;1:602–609. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous