The histone deacetylase HDAC3 targets RbAp48 to the retinoblastoma protein - PubMed (original) (raw)
The histone deacetylase HDAC3 targets RbAp48 to the retinoblastoma protein
E Nicolas et al. Nucleic Acids Res. 2001.
Abstract
The product of the retinoblastoma susceptibility gene, the Rb protein, functions partly through transcriptional repression of E2F-regulated genes. Repression by Rb is mediated, at least in part, by a histone deacetylase complex, whose enzymatic activity relies on HDAC1, HDAC2 or HDAC3. Recently, we have shown that the Rb-associated histone deacetylase complex contains RbAp48 protein, which interacts with HDAC1 and HDAC2. RbAp48 could favour the deacetylation of histones since it binds directly to histone H4. In agreement with that, we show that transcriptional repression of E2F activity requires the presence of RbAp48. HDAC3 was thought not to interact with RbAp48. However, we found that it shared with HDAC1 the ability to favour the recruitment of RbAp48 to Rb. This latter effect was unlikely to be due to activation of Rb function, since HDAC3 did not increase Rb-E2F1 interaction. Rather, we found, surprisingly, that HDAC3 could physically interact with RbAp48 both in vitro and in living cells. Taken together, our data suggest a model in which Rb mediates the recruitment to E2F-regulating promoters of a repressive complex containing either HDAC1, HDAC2 or HDAC3 and the histone-binding protein RbAp48.
Figures
Figure 1
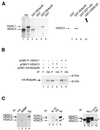
Exogenous HDAC3 increases the Rb–RbAp48 interaction. SAOS-2 cells were transfected with 5 µg of the indicated expression vectors [the Rb expression vector encodes the Rb pocket domain (379–928)] by the calcium phosphate co-precipitation method. The amount of promoter in the transfection was kept constant using empty vectors. Twenty-four hours after transfection, total cell lysates were prepared and immunoprecipitated as described (16), using 1 µg of the indicated antibody [anti-Rb, antibody C15G (Santa Cruz Biotechnologies); anti-HA, antibody 12CA5 (Roche Diagnostics)]. Immunoprecipitates were subjected to western blot analysis using the anti-HA antibody (top) or an anti-Rb antibody (antibody XZ55; Pharmingen) (bottom) using standard procedures. The arrows indicate the position of the exogenous proteins. Note that exogenous Rb migrates at ∼60 kDa, because the expression vector used in these experiments expressed a version of Rb deleted for the first 378 amino acids of the molecule.
Figure 2
Exogenous HDAC3 does not increase the E2F1–Rb interaction. SAOS-2 cells were transiently transfected by calcium phosphate co-precipitation with the indicated expression vectors. Total cell lysates were immunoprecipitated using the anti-Rb antibody (top and middle) or an anti-E2F1 antibody (antibody KH95; Santa Cruz Biotechnologies) (bottom). Immunoprecipitates were subjected to western blot analysis using the anti-E2F1 antibody (top and bottom) or the anti-Rb antibody (middle).
Figure 3
Physical association between HDAC3 and RbAp48. (A) 35S-labelled in vitro translated HDAC1 (lanes 5 and 6) or HDAC3 (lanes 3–4 and 7–10) was subjected to GST pull down analysis using beads harbouring 1 µg GST–RbAp48 fusion protein (lanes 3, 5 and 8), control GST (lanes 4, 6 and 7), GST–E2F1 AD (lane 9) or GST–CREB AD (lane 10), as indicated. After extensive washing, bound proteins were analysed by SDS–PAGE followed by autoradiography. In lanes 1 and 2, 10% of the amount of in vitro translated HDAC3 or HDAC1 used in the pull down reaction was directly loaded. (B) SAOS-2 cells were transiently transfected by calcium phosphate co-precipitation with the indicated expression vector and total cell extracts were immunoprecipitated with the indicated antibody (anti-Flag M2 antibody, antibody F; Sigma). Immunoprecipitates were subjected to western blot analysis using the anti-HA antibody. The arrows indicate the position of the two RbAp48 bands. (C) Hela nuclear extracts [50 (lanes 1, 2, 6 and 7) or 200 µl (lanes 11–12)] were immunoprecipitated with 1 µg of either an anti-RbAp48 antibody [lane 2, antibody RBBP (Transduction Laboratories); lane 6, antibody N19 (Santa-Cruz); lane 11, antibody 11G10 (Genetex)] or control anti-HA antibody [lanes 1, 7 and 12, antibody 12CA5 (Roche Diagnostics)]. In lanes 4, 8 and 10, 4 µl of HeLa nuclear extracts were directly loaded. In lanes 3, 5 and 9, 1 µg RBBP, N19 and 11G10 antibodies, respectively, were loaded, to monitor the migration of immunoglobulins. Immunoprecipitates were tested for the presence of HDAC1, HDAC2 and HDAC3 by western blotting using an anti-HDAC antibody (Transduction Laboratories). The stars indicate bands due to the immunoglobulin heavy chains from the anti-RbAp48 antibody (lanes 9 and 11) or the anti-HA antibody (lanes 7 and 12). Note that at longer exposures HDAC1 and HDAC2 could be detected in RBBP immunoprecipitates (lane 2). Also, the amount of HDAC3 in N-19 immunoprecipitates (lane 6) is likely to be overestimated due to co-migration with the immunoglobulin heavy chains, which were weakly detected (lane 5).
Figure 4
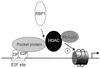
Model of transcriptional repression by Rb through the recruitment of histone deacetylases. In G0 or during the beginning of the G1 phase of the cell cycle Rb protein (or one of its cousins, ‘Pocket protein’ in the figure) binds to E2F sites (15). It recruits a histone deacetylase (‘HDAC’) through a direct (HDAC1 or HDAC2) or indirect (HDAC3, through RBP1) interaction. These three deacetylases share the ability to recruit the histone-binding protein RbAp48, leading to deacetylation of histones present on the promoter.
Similar articles
- RbAp48 belongs to the histone deacetylase complex that associates with the retinoblastoma protein.
Nicolas E, Morales V, Magnaghi-Jaulin L, Harel-Bellan A, Richard-Foy H, Trouche D. Nicolas E, et al. J Biol Chem. 2000 Mar 31;275(13):9797-804. doi: 10.1074/jbc.275.13.9797. J Biol Chem. 2000. PMID: 10734134 - Functional and physical interaction between the histone methyl transferase Suv39H1 and histone deacetylases.
Vaute O, Nicolas E, Vandel L, Trouche D. Vaute O, et al. Nucleic Acids Res. 2002 Jan 15;30(2):475-81. doi: 10.1093/nar/30.2.475. Nucleic Acids Res. 2002. PMID: 11788710 Free PMC article. - Repression of E2F1-mediated transcription by the ErbB3 binding protein Ebp1 involves histone deacetylases.
Zhang Y, Woodford N, Xia X, Hamburger AW. Zhang Y, et al. Nucleic Acids Res. 2003 Apr 15;31(8):2168-77. doi: 10.1093/nar/gkg318. Nucleic Acids Res. 2003. PMID: 12682367 Free PMC article. - Transcriptional control at regulatory checkpoints by histone deacetylases: molecular connections between cancer and chromatin.
Wade PA. Wade PA. Hum Mol Genet. 2001 Apr;10(7):693-8. doi: 10.1093/hmg/10.7.693. Hum Mol Genet. 2001. PMID: 11257101 Review. - Small molecule regulators of Rb-E2F pathway as modulators of transcription.
Singh S, Johnson J, Chellappan S. Singh S, et al. Biochim Biophys Acta. 2010 Oct-Dec;1799(10-12):788-94. doi: 10.1016/j.bbagrm.2010.07.004. Epub 2010 Jul 15. Biochim Biophys Acta. 2010. PMID: 20637913 Free PMC article. Review.
Cited by
- Comparative Review on Cancer Pathology from Aberrant Histone Chaperone Activity.
Lee J, Bao X. Lee J, et al. Int J Mol Sci. 2024 Jun 10;25(12):6403. doi: 10.3390/ijms25126403. Int J Mol Sci. 2024. PMID: 38928110 Free PMC article. Review. - RebL1 is required for macronuclear structure stability and gametogenesis in Tetrahymena thermophila.
Hao H, Lian Y, Ren C, Yang S, Zhao M, Bo T, Xu J, Wang W. Hao H, et al. Mar Life Sci Technol. 2024 Mar 26;6(2):183-197. doi: 10.1007/s42995-024-00219-z. eCollection 2024 May. Mar Life Sci Technol. 2024. PMID: 38827131 Free PMC article. - Acetyl-methyllysine marks chromatin at active transcription start sites.
Lu-Culligan WJ, Connor LJ, Xie Y, Ekundayo BE, Rose BT, Machyna M, Pintado-Urbanc AP, Zimmer JT, Vock IW, Bhanu NV, King MC, Garcia BA, Bleichert F, Simon MD. Lu-Culligan WJ, et al. Nature. 2023 Oct;622(7981):173-179. doi: 10.1038/s41586-023-06565-9. Epub 2023 Sep 20. Nature. 2023. PMID: 37731000 Free PMC article. - Understanding the Role of Histone Deacetylase and their Inhibitors in Neurodegenerative Disorders: Current Targets and Future Perspective.
Kumar V, Kundu S, Singh A, Singh S. Kumar V, et al. Curr Neuropharmacol. 2022;20(1):158-178. doi: 10.2174/1570159X19666210609160017. Curr Neuropharmacol. 2022. PMID: 34151764 Free PMC article. Review. - Anticancer potential of the histone deacetylase inhibitor-like effects of flavones, a subclass of polyphenolic compounds: a review.
Singh P, Tomar RS, Rath SK. Singh P, et al. Mol Biol Rep. 2015 Nov;42(11):1515-31. doi: 10.1007/s11033-015-3881-y. Epub 2015 Jun 2. Mol Biol Rep. 2015. PMID: 26033434 Review.
References
- Muller H. and Helin,K. (2000) The E2F transcription factors: key regulators of cell proliferation. Biochim. Biophys. Acta, 1470, M1–M12. - PubMed
- Black A.R. and Azizkhan-Clifford,J. (1999) Regulation of E2F: a family of transcription factors involved in proliferation control. Gene, 237, 281–302. - PubMed
- Kaelin W.G.Jr (1999) Functions of the retinoblastoma protein. Bioessays, 21, 950–958. - PubMed
- Krek W., Xu,G. and Livingston,D.M. (1995) Cyclin A-kinase regulation of E2F-1 DNA binding function underlies suppression of an S phase checkpoint. Cell, 83, 1149–1158. - PubMed
- Zhang H.S., Postigo,A.A. and Dean,D.C. (1999) Active transcriptional repression by the Rb-E2F complex mediates G1 arrest triggered by p16INK4a, TGFbeta and contact inhibition. Cell, 97, 53–61. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous