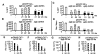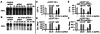Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems - PubMed (original) (raw)
Comparative Study
. 2001 Aug 14;98(17):9742-7.
doi: 10.1073/pnas.171251798. Epub 2001 Jul 31.
Affiliations
- PMID: 11481446
- PMCID: PMC55523
- DOI: 10.1073/pnas.171251798
Comparative Study
Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems
N J Caplen et al. Proc Natl Acad Sci U S A. 2001.
Abstract
Short interfering RNAs (siRNAs) are double-stranded RNAs of approximately 21-25 nucleotides that have been shown to function as key intermediaries in triggering sequence-specific RNA degradation during posttranscriptional gene silencing in plants and RNA interference in invertebrates. siRNAs have a characteristic structure, with 5'-phosphate/3'-hydroxyl ends and a 2-base 3' overhang on each strand of the duplex. In this study, we present data that synthetic siRNAs can induce gene-specific inhibition of expression in Caenorhabditis elegans and in cell lines from humans and mice. In each case, the interference by siRNAs was superior to the inhibition of gene expression mediated by single-stranded antisense oligonucleotides. The siRNAs seem to avoid the well documented nonspecific effects triggered by longer double-stranded RNAs in mammalian cells. These observations may open a path toward the use of siRNAs as a reverse genetic and therapeutic tool in mammalian cells.
Figures
Figure 1
Gene-specific inhibition of expression in MEFs by siRNAs. MEFs transfected with plasmid DNA, ssRNAs, and dsRNAs were harvested 48 h after transfection and were assayed for (A_–_D) GFP expression by FACS analysis (each transfection was assayed in triplicate and data are shown as mean ± SEM). A and C show the percentage of GFP-positive cells and B and_D_ show the fluorescence intensity (Geo Mean) of GFP-positive cells. (E_–_I) CAT expression (each transfection condition was assayed in triplicate; data in_E_ are normalized to the amount of CAT pg/μg of protein observed in pcDNA3.CAT-transfected cells; data in_F_–I are normalized to the amount of CAT pg/μg of protein in plasmid and sense ssRNA-transfected cells. s, sense ssRNA; as, antisense ssRNA). (J)egfp and neo RNA levels by Northern analysis of poly(A)+ mRNA. (K) Cell survival (assayed in duplicate and shown as a mean OD560–650; dsRNAs of 21–25 and 78 nucleotides correspond to egfp; the dsRNA of 81 nucleotides corresponds to LacZ). (L and M) GFP expression by FACS analysis (data are shown as relative percentage normalized to pEGFP-N3-transfected cells). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Figure 2
siRNA-mediated gene silencing in human cells. (A and_B_) 293 and (C and D) HeLa cells transfected with pEGFP-N3 and antisense (as) ssRNAs and dsRNAs were harvested 48 h after transfection and were assayed for GFP expression by FACS analysis (assayed in triplicate; data are shown as mean ± SEM). A and C show the percentage of GFP-positive cells and B and_D_ show the fluorescence intensity (Geo Mean) of GFP-positive cells. (E_–_H) HeLa cells transfected with pcDNA3-CAT, ssRNAs, and dsRNAs were harvested 48 h after transfection and assayed for CAT expression (assayed in triplicate and normalized to the amount of CAT pg/μg of protein observed in plasmid plus sense-transfected cells. s, sense ssRNA; as, antisense ssRNA). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Figure 3
siRNAs and mammalian dsRNA-dependent pathways. To detect PKR autophosphorylation, we performed in vitro kinase assays as described in Methods. (A) In vitro kinase reactions were performed without exogenous RNA (−) or with 1 μg/ml of reovirus dsRNA or 1 μg/ml of siRNA (21–25 nucleotides), or 1 μg/ml of 78- or 81-nt dsRNA. (B) In vitro kinase competition assays were performed by using si- and dsRNAs. Reactions were performed without exogenous RNA (−) or 1 μg/ml of reovirus RNA, or 75-fold excess siRNA (21–25 nucleotides) or 78- or 81-nt dsRNA, plus reovirus dsRNA (1 μg/ml). siRNAs of 21–25 nucleotides and dsRNA of 78 nucleotides corresponded to egfp (the 81-nt dsRNA corresponds to LacZ). (C and_D_) 293 cells transfected with pEGFP-N3 and_unc-22_ or egfp siRNAs, and (E and F) 293 cells transfected with pEGFP-N3 and 78 egfp dsRNA and unc-22 or_egfp_ siRNAs were assayed for GFP expression by FACS analysis 48 h after transfection (each transfection was assayed in triplicate; data are shown as mean ± SEM). B and_D_ show the percentage of GFP-positive cells and_C_ and E show the fluorescence intensity (Geo Mean) of GFP-positive cells. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Similar articles
- On the role of RNA amplification in dsRNA-triggered gene silencing.
Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH, Fire A. Sijen T, et al. Cell. 2001 Nov 16;107(4):465-76. doi: 10.1016/s0092-8674(01)00576-1. Cell. 2001. PMID: 11719187 - Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans.
Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Fire A, et al. Nature. 1998 Feb 19;391(6669):806-11. doi: 10.1038/35888. Nature. 1998. PMID: 9486653 - Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines.
Billy E, Brondani V, Zhang H, Müller U, Filipowicz W. Billy E, et al. Proc Natl Acad Sci U S A. 2001 Dec 4;98(25):14428-33. doi: 10.1073/pnas.261562698. Epub 2001 Nov 27. Proc Natl Acad Sci U S A. 2001. PMID: 11724966 Free PMC article. - Gene silencing: shrinking the black box of RNAi.
Hunter CP. Hunter CP. Curr Biol. 2000 Feb 24;10(4):R137-40. doi: 10.1016/s0960-9822(00)00325-0. Curr Biol. 2000. PMID: 10704407 Review. - Small RNA: can RNA interference be exploited for therapy?
Wall NR, Shi Y. Wall NR, et al. Lancet. 2003 Oct 25;362(9393):1401-3. doi: 10.1016/S0140-6736(03)14637-5. Lancet. 2003. PMID: 14585643 Review.
Cited by
- Gene editing of angiotensin for blood pressure management.
Masi S, Dalpiaz H, Borghi C. Masi S, et al. Int J Cardiol Cardiovasc Risk Prev. 2024 Aug 20;23:200323. doi: 10.1016/j.ijcrp.2024.200323. eCollection 2024 Dec. Int J Cardiol Cardiovasc Risk Prev. 2024. PMID: 39258007 Free PMC article. - RNA Interference based Midkine Gene Therapy for Hepatocellular Carcinoma.
Mamdouh S, Khorshed FEM, Hammad G, Magdy M, Abdelraouf A, Hemida E, Shemis M. Mamdouh S, et al. Asian Pac J Cancer Prev. 2024 Jul 1;25(7):2371-2379. doi: 10.31557/APJCP.2024.25.7.2371. Asian Pac J Cancer Prev. 2024. PMID: 39068570 Free PMC article. - RNA-Based Antipsoriatic Gene Therapy: An Updated Review Focusing on Evidence from Animal Models.
Lin ZC, Hung CF, Aljuffali IA, Lin MH, Fang JY. Lin ZC, et al. Drug Des Devel Ther. 2024 Apr 23;18:1277-1296. doi: 10.2147/DDDT.S447780. eCollection 2024. Drug Des Devel Ther. 2024. PMID: 38681207 Free PMC article. Review. - siRNA Therapeutics: From Bench Lab. to Clinics.
Romero-López C, Berzal-Herranz A. Romero-López C, et al. Pharmaceuticals (Basel). 2024 Mar 26;17(4):416. doi: 10.3390/ph17040416. Pharmaceuticals (Basel). 2024. PMID: 38675378 Free PMC article. - RNA therapeutics to control fibrinolysis: review on applications in biology and medicine.
Ferraresso F, Leung J, Kastrup CJ. Ferraresso F, et al. J Thromb Haemost. 2024 Aug;22(8):2103-2114. doi: 10.1016/j.jtha.2024.04.006. Epub 2024 Apr 24. J Thromb Haemost. 2024. PMID: 38663489 Review.
References
- Wolffe A P, Matzke M A. Science. 1999;286:481–486. - PubMed
- Frischmeyer P A, Dietz H C. Hum Mol Genet. 1999;8:1893–1900. - PubMed
- Mitchell P, Tollervey D. Curr Opin Genet Dev. 2000;10:193–198. - PubMed
- Fire A. Trends Genet. 1999;15:358–363. - PubMed
- Cogoni C, Macino G. Curr Opin Genet Dev. 2000;10:638–643. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM037706/GM/NIGMS NIH HHS/United States
- AF R01-GM37706/GM/NIGMS NIH HHS/United States
- FI AI446962/AI/NIAID NIH HHS/United States
- T32-GM07321/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources