Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells - PubMed (original) (raw)
Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells
A Loonstra et al. Proc Natl Acad Sci U S A. 2001.
Abstract
The use of Cre/loxP recombination in mammalian cells has expanded rapidly. We describe here that Cre expression in cultured mammalian cells may result in a markedly reduced proliferation and that this effect is dependent on the endonuclease activity of Cre. Chromosome analysis after Cre expression revealed numerous chromosomal aberrations and an increased number of sister chromatid exchanges. Titration experiments in mouse embryo fibroblasts with a ligand-regulatable Cre-ER(T) show that toxicity is dependent on the level of Cre activity. Prolonged, low levels of Cre activity permit recombination without concomitant toxicity. This urges for a careful titration of Cre activity in conditional gene modification in mammalian cells.
Figures
Figure 1
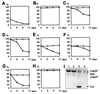
Repressive effects of Cre endonuclease activity on proliferation of mammalian cells. (A) Selective growth repression of MEFs infected with bicistronic retroviruses expressing Cre linked to GFP. Every 72 h, cells were enumerated before replating and analyzed by flow cytometry. (B) Growth rates of MEFs expressing GFP alone are comparable to wt cells. Primary MEFs (C), NIH 3T3 (D), COS-7 (E), HeLa (F), and U2OS (G) cells infected with bicistronic retroviruses expressing Cre-ERT and GFP show a reduced proliferation upon culturing in the presence of 1 μM OHT. No difference in proliferation is observed between wt cells and infected cells in the absence of OHT. (H) MEFs infected with retroviruses encoding an endonuclease-deficient Cre-ERT fusion protein do not suffer from a Cre-induced proliferation defect when cultured in the presence of OHT. The curves representing growth in the presence and absence of OHT are indistinguishable. For B_–_H, the data points are as follows: ○, cultured without OHT; ●, cultured in presence of 1 μM OHT. (I) Western blot analysis of expression levels of Cre and Cre fusion proteins in MEFs infected with viruses encoding Cre-ERT (lane 1), Cre(R173K)-ERT (lane 2), and Cre (lane 3), and in _R26cre-ER_T MEFs (lane 4). The R26cre-ERT fusion protein is slightly shorter than the retrovirally encoded Cre-ERT and Cre(R173K)-ERT fusion proteins because of a 21-aa deletion in domain D of ERT (29).
Figure 2
Characteristics of Cre-mediated recombination and growth inhibition in _R26cre-ER_T MEFs. (A and B) Time and dose dependence of Cre-ERT-mediated recombination. (A) Southern blot analysis of DNA from _Brca2_F11F/wt; _R26cre-ER_T MEFs that were treated with 1 μM OHT for the indicated periods of time. Control DNA from _Brca2_Δ/wt cells is included (lane C). (B) Southern blot analysis of DNA from _Brca2_F11F/wt; _R26cre-ER_T MEFs that were treated with various concentrations of OHT for 24 h. After OHT treatment, cells were cultured for another 24 h to allow for clearance of OHT-bound Cre-ERT. Hybridization signals indicate the floxed exon 11 or wt alleles (f/wt) or the deleted exon 11 allele (Δ) of Brca2. (C) Growth inhibition in _R26cre-ER_T MEFs after treatment with 1 μM OHT. Cells were plated at 2 × 105 per 10-cm2 dish and cultured in medium with 1 μM OHT for 0 h (●), 24 h (○), or 48 h (▴). On day 3 of the experiment, cells were counted, replated at initial densities, and cultured for another 3 days. The growth curves represent cumulative cell numbers. (D) Kinetic analysis for growth of R26 Cre-ERT MEFs in the presence of 0 μM (●), 0.1 μM (○), 0.3 μM (▴), or 1 μM (■) of OHT. (E and F) Prolonged, low-level expression of Cre results in detectable recombination without growth inhibition. (E) Growth curves of _Brca2_F11F/wt; _R26cre-ER_T MEFs cultured in the presence of 0 μM (●), 0.05 μM (■), 0.1 μM (▴), 0.2 μM (○), 0.3 μM (□), or 0.6 μM (▵) of OHT. Cells were plated at 2 × 1055 per 10-cm2 dish, and every 3 days cells were enumerated before replating. The growth curves represent cumulative cell numbers. (F) After 3 and 9 days of OHT treatment, DNA from the cell cultures described in E was used to detect deletion of the floxed Brca2 exon 11 by Southern blotting. OHT concentrations are indicated above the panes; recombination percentages, below.
Figure 3
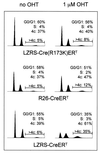
Cre expression in MEFs induces accumulation in G2/M and aneuploidy. Cell cycle analysis of MEFs infected with viruses encoding Cre-ERT, Cre(R173K)-ERT, Cre (lane 3), and _R26cre-ER_T MEFs, either untreated or treated with 1 μM OHT, for 3 days. Cells were stained with propidium iodide for DNA content and analyzed by FACS. The numbers indicate the percentage of nonpolyploid cells in G0/G1, S, and G2/M phases of the cell cycle. In addition, a region marker is set to determine the percentage of polyploid cells in the total population.
Figure 4
Cre expression in MEFs induces chromosome abnormalities and increased frequencies of SCE. (A) Expression of active Cre induces chromosome aberrations in MEFs. Metaphase preparations of wt or _R26cre-ER_T MEFs either untreated or treated with 1 μM OHT for 48 h were examined by eye for the presence of chromosomal aberrations. n, number of metaphases examined. (B_–_D) SKY analysis of a typical metaphase from _R26cre-ER_T MEFs treated for 48 h with 1 μM OHT. (B) An individual metaphase after hybridization with fluorescent probes. The 4′,6-diamidino-2-phenylindole-banded image of the same metaphase is shown in C and in display colors by assignation of hybridization signals to specific spectral ranges (D). The arrowheads in B and C indicate examples of dicentric and acentric chromosome fusions and fragmented chromosomes. (D) Multiple fragmented chromosomes (2, 4, 7, 8, 12, and 16), two acentric fusions of chromosome 2 and one of chromosome 5, and dicentric fusions of chromosomes 4 and 14, chromosomes 14 and 18, and chromosomes X. (E) Expression of active Cre causes increased SCE in MEFs. Metaphase preparations of wt or _R26cre-ER_T MEFs either untreated or treated with 1 μM OHT for 48 h were labeled with BrdUrd before preparation of metaphase spreads. The number of SCEs per cell was determined for at least 40 diploid metaphases per culture.
Similar articles
- Cre recombinase induces DNA damage and tetraploidy in the absence of loxP sites.
Janbandhu VC, Moik D, Fässler R. Janbandhu VC, et al. Cell Cycle. 2014;13(3):462-70. doi: 10.4161/cc.27271. Epub 2013 Nov 26. Cell Cycle. 2014. PMID: 24280829 Free PMC article. - Efficient Cre-loxP-induced mitotic recombination in mouse embryonic stem cells.
Liu P, Jenkins NA, Copeland NG. Liu P, et al. Nat Genet. 2002 Jan;30(1):66-72. doi: 10.1038/ng788. Epub 2001 Dec 10. Nat Genet. 2002. PMID: 11740496 - Induced DNA recombination by Cre recombinase protein transduction.
Joshi SK, Hashimoto K, Koni PA. Joshi SK, et al. Genesis. 2002 May;33(1):48-54. doi: 10.1002/gene.10089. Genesis. 2002. PMID: 12001069 - Cre-loxP biochemistry.
Ghosh K, Van Duyne GD. Ghosh K, et al. Methods. 2002 Nov;28(3):374-83. doi: 10.1016/s1046-2023(02)00244-x. Methods. 2002. PMID: 12431441 Review. - Cre Recombinase.
Van Duyne GD. Van Duyne GD. Microbiol Spectr. 2015 Feb;3(1):MDNA3-0014-2014. doi: 10.1128/microbiolspec.MDNA3-0014-2014. Microbiol Spectr. 2015. PMID: 26104563 Review.
Cited by
- BEAM: A combinatorial recombinase toolbox for binary gene expression and mosaic genetic analysis.
Greig LC, Woodworth MB, Poulopoulos A, Lim S, Macklis JD. Greig LC, et al. Cell Rep. 2024 Aug 27;43(8):114650. doi: 10.1016/j.celrep.2024.114650. Epub 2024 Aug 17. Cell Rep. 2024. PMID: 39159043 Free PMC article. - NatB Protects Procaspase-8 from UBR4-Mediated Degradation and Is Required for Full Induction of the Extrinsic Apoptosis Pathway.
Guedes JP, Boyer JB, Elurbide J, Carte B, Redeker V, Sago L, Meinnel T, Côrte-Real M, Giglione C, Aldabe R. Guedes JP, et al. Mol Cell Biol. 2024;44(9):358-371. doi: 10.1080/10985549.2024.2382453. Epub 2024 Aug 4. Mol Cell Biol. 2024. PMID: 39099191 - Mechanisms underlying the direct programming of mouse embryonic fibroblasts to thymic epithelial cells by FOXN1.
Ma Z, Kang S, Condie BG, Manley NR. Ma Z, et al. Development. 2024 Jul 15;151(14):dev202730. doi: 10.1242/dev.202730. Epub 2024 Jul 22. Development. 2024. PMID: 38958026 - How Much Do You Fuse? A Comparison of Cell Fusion Assays in a Breast Cancer Model.
Sieler M, Dörnen J, Dittmar T. Sieler M, et al. Int J Mol Sci. 2024 May 23;25(11):5668. doi: 10.3390/ijms25115668. Int J Mol Sci. 2024. PMID: 38891857 Free PMC article. - iSuRe-HadCre is an essential tool for effective conditional genetics.
Garcia-Gonzalez I, Rocha SF, Hamidi A, Garcia-Ortega L, Regano A, Sanchez-Muñoz MS, Lytvyn M, Garcia-Cabero A, Roig-Soucase S, Schoofs H, Castro M, Sabata H, Potente M, Graupera M, Makinen T, Benedito R. Garcia-Gonzalez I, et al. Nucleic Acids Res. 2024 Jul 22;52(13):e56. doi: 10.1093/nar/gkae472. Nucleic Acids Res. 2024. PMID: 38850155 Free PMC article.
References
- Sternberg N, Hamilton D. J Mol Biol. 1981;150:467–486. - PubMed
- Sternberg N, Hamilton D, Austin S, Yarmolinsky M, Hoess R. Cold Spring Harbor Symp Quant Biol. 1981;45:297–309. - PubMed
- Abremski K, Hoess R. J Biol Chem. 1984;259:1509–1514. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources