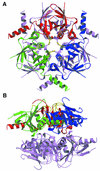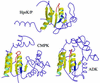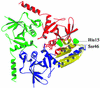X-ray structure of HPr kinase: a bacterial protein kinase with a P-loop nucleotide-binding domain - PubMed (original) (raw)
X-ray structure of HPr kinase: a bacterial protein kinase with a P-loop nucleotide-binding domain
S Fieulaine et al. EMBO J. 2001.
Abstract
HPr kinase/phosphatase (HprK/P) is a key regulatory enzyme controlling carbon metabolism in Gram- positive bacteria. It catalyses the ATP-dependent phosphorylation of Ser46 in HPr, a protein of the phosphotransferase system, and also its dephosphorylation. HprK/P is unrelated to eukaryotic protein kinases, but contains the Walker motif A characteristic of nucleotide-binding proteins. We report here the X-ray structure of an active fragment of Lactobacillus casei HprK/P at 2.8 A resolution, solved by the multiwavelength anomalous dispersion method on a seleniated protein (PDB code 1jb1). The protein is a hexamer, with each subunit containing an ATP-binding domain similar to nucleoside/nucleotide kinases, and a putative HPr-binding domain unrelated to the substrate-binding domains of other kinases. The Walker motif A forms a typical P-loop which binds inorganic phosphate in the crystal. We modelled ATP binding by comparison with adenylate kinase, and designed a tentative model of the complex with HPr based on a docking simulation. The results confirm that HprK/P represents a new family of protein kinases, first identified in bacteria, but which may also have members in eukaryotes.
Figures
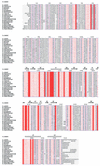
Fig. 1. Alignment of bacterial HprK/P sequences. The secondary structure on top is defined by DSSP (Kabsch and Sander, 1983) for the truncated L.casei subunit. Blue frames are for conserved residues, white characters in red boxes for strict identity, and red characters in white boxes for similarity. Accession numbers for Gram-positive bacteria: Lactobacillus casei SwissProt Q9RE09, Enterococcus faecalis SwissProt O07664, Streptococcus mutans SwissProt Q9ZA56, Streptococcus bovis SwissProt Q9WXK7, Streptococcus pyogenes orf1717 (
http://pedant.mips.biochem.mpg.de
), Streptococcus salivarius SwissProt Q9ZA98, Lactococcus lactis MOLOKO database (
), Bacillus subtilis Swiss-Prot O34483, Bacillus halodurans TrEMBL P82557, Staphylococcus xylosus SwissProt Q9S1H5, Clostridium acetobutylicum (
http://www.genomecorp.com/htdocs/sequences/clostridium/clospage.html
), Mycoplasma genitalium SwissProt P47331, Mycoplasma pneumoniae Swiss-Prot P75548, Ureaplasma parvum SwissProt Q9PR69. Gram-negative bacteria: Treponema pallidum SwissProt O83600, Neisseria gonorrhoeae (
http://www.ncbi.nlm.nih.gov/Microb\_blast/unfinishedgenome.html
), Neisseria meningitidis TrEMBL Q9K6Y4, Xylella fastidiosa SwissProt Q9PDH3. Figure realized with ESPript (Gouet et al., 1999).
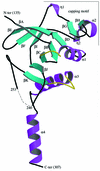
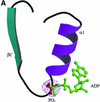
Fig. 2. The L.casei HprK/P fold: a ribbon diagram of the truncated 128–319 monomer. The N- and C-terminal residues and residues 241–252 (dashes) are disordered and missing from the model. The monomer contains 11 β-strands βA–βK and four α-helices α1–α4. The ‘capping motif’ on top includes βG, βH, α2 and two single-turn 310-helices labelled η1 and η2. The P-loop (residues 155–160), linking α1 to βC, and the K3 loop (residues 267–272), linking βK to α3, are highlighted in yellow. The P-loop contains the Walker A motif and the phosphate-binding site; the K3 loop is involved in trimer contacts. This and the following figures were drawn using MOLSCRIPT (Esnouf, 1997) and RASTER 3D (Merritt and Bacon, 1997), except Figures 4B and 6A drawn with BOBSCRIPT (Esnouf, 1999).
Fig. 3. The HprK/P hexamer. The hexamer forms a 55 Å thick two-layered structure with a trimer of 80 Å diameter in each layer. The bottom trimer is in mauve. Each subunit of the top trimer is in a different colour, with the P-loop and the K3 loop highlighted in yellow. (A) View along the 3-fold axis. The six K3 loops are at the centre, the N- and C-terminal ends at the periphery of the hexamer. (B) Orthogonal view along the 2-fold axis. The P-loops, K3 loops and the disordered 241–252 residues are all located at the surface of the trimers.
Fig. 4. Subunit contacts in the hexamer. (A) The trimer. Regions involved in the contacts are highlighted in red, green and blue, respectively, for each subunit. C-terminal helices α3 and α4 from the blue subunit pack against helix α1 and the capping motif of the red subunit. Additional contacts involve the K3 and P-loops in yellow. (B) Dimer contacts. One subunit of the bottom trimer makes contact with all three subunits of the top trimer, mostly through loops at the edge of the central β-sheet, the C-terminus of the capping motif and the C-terminus of helix α3. On the 2-fold axis, density in a (2_F_o – _F_c) map countoured at 1σ is interpreted as a set of water molecules. It bridges His173 from the bottom and top subunits.
Fig. 5. Comparison of HprK/P with cytidylate and adenylate kinases. The conserved α/β structural motif is highlighted in yellow ribbon in the HprK/P subunit and the two proteins with the highest DALI scores, E.coli cytidine monophosphate kinase (CMPK, PDB code 1cke) and B.stearothermophilus adenylate kinase (ADK, PDB code 1zin). The conserved nucleotide-binding motif includes a five-stranded parallel β-sheet and two α-helices (with the P-loop highlighted in red). However, in HprK/P, βB (in blue) is antiparallel to the other four strands whilst all strands in CMPK and ADK are parallel and the topology differs.
Fig. 6. The P-loop and bound phosphate. (A) The P-loop connecting strand βC to helix α1. A bound phosphate ion (in red) is shown in its 2_F_o – _F_c electron density map contoured at 1σ. The ADP molecule in green is positioned by comparison with Sulfolobus acidocaldarius adenylate kinase (PDB code 1nks-F) (Vonrhein et al., 1998). Its β-phosphate overlaps the phosphate ion of HprK/P. (B) Stereo view of interactions made by the phosphate ion. Hydrogen bonds (dashes) link oxygen atoms of the phosphate ion to the four NH groups of P-loop residues 158–162 and to the side chains of Lys161, Ser162 and Glu163.
Fig. 6. The P-loop and bound phosphate. (A) The P-loop connecting strand βC to helix α1. A bound phosphate ion (in red) is shown in its 2_F_o – _F_c electron density map contoured at 1σ. The ADP molecule in green is positioned by comparison with Sulfolobus acidocaldarius adenylate kinase (PDB code 1nks-F) (Vonrhein et al., 1998). Its β-phosphate overlaps the phosphate ion of HprK/P. (B) Stereo view of interactions made by the phosphate ion. Hydrogen bonds (dashes) link oxygen atoms of the phosphate ion to the four NH groups of P-loop residues 158–162 and to the side chains of Lys161, Ser162 and Glu163.
Fig. 7. A model of the interaction of HPr with HprK/P. The serine-phosphorylated form of E.faecalis HPr (Audette et al., 2000) is drawn in yellow in the position and orientation determined by the docking simulation described in the text. Green van der Waals spheres represent the phosphorylated Ser46 and phosphorylable His15 side chains. HPr is predicted to sit on the top of a HprK/P trimer and interact with both the red and the blue subunits. Its phosphoserine is near the P-loop of the red subunit, which also interacts with HPr via its capping motif. The C-terminal helix α4 of the blue subunit sits in between αA and αC of HPr. An ADP molecule is drawn in ball-and-stick bound to the red subunit as described in Figure 6.
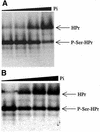
Fig. 8. Activity assays with truncated L.casei HprK/P. (A) HPr kinase assay. HPr at 12 µM was incubated for 10 min at 37°C with 400 nM HprK/P and increasing concentrations of Pi (0, 2, 5 and 20 mM) in the presence of 5 mM MgCl2, 10 mM FBP and 1 mM ATP. (B) P-Ser-HPr phosphatase assay. P-Ser-HPr at 12 µM was incubated for 5 min at 37°C with 88 nM HprK/P and increasing concentrations of Pi (0, 0.2, 1 and 3 mM) in the presence of 5 mM MgCl2.
Similar articles
- X-ray structure of a bifunctional protein kinase in complex with its protein substrate HPr.
Fieulaine S, Morera S, Poncet S, Mijakovic I, Galinier A, Janin J, Deutscher J, Nessler S. Fieulaine S, et al. Proc Natl Acad Sci U S A. 2002 Oct 15;99(21):13437-41. doi: 10.1073/pnas.192368699. Epub 2002 Oct 1. Proc Natl Acad Sci U S A. 2002. PMID: 12359875 Free PMC article. - Crystal structure of HPr kinase/phosphatase from Mycoplasma pneumoniae.
Allen GS, Steinhauer K, Hillen W, Stülke J, Brennan RG. Allen GS, et al. J Mol Biol. 2003 Feb 28;326(4):1203-17. doi: 10.1016/s0022-2836(02)01378-5. J Mol Biol. 2003. PMID: 12589763 - Structural analysis of the bacterial HPr kinase/phosphorylase V267F mutant gives insights into the allosteric regulation mechanism of this bifunctional enzyme.
Chaptal V, Vincent F, Gueguen-Chaignon V, Monedero V, Poncet S, Deutscher J, Nessler S, Morera S. Chaptal V, et al. J Biol Chem. 2007 Nov 30;282(48):34952-7. doi: 10.1074/jbc.M705979200. Epub 2007 Sep 18. J Biol Chem. 2007. PMID: 17878158 - HPr kinase/phosphorylase, a Walker motif A-containing bifunctional sensor enzyme controlling catabolite repression in Gram-positive bacteria.
Poncet S, Mijakovic I, Nessler S, Gueguen-Chaignon V, Chaptal V, Galinier A, Boël G, Mazé A, Deutscher J. Poncet S, et al. Biochim Biophys Acta. 2004 Mar 11;1697(1-2):123-35. doi: 10.1016/j.bbapap.2003.11.018. Biochim Biophys Acta. 2004. PMID: 15023355 Review. - Transcription regulators potentially controlled by HPr kinase/phosphorylase in Gram-negative bacteria.
Boël G, Mijakovic I, Mazé A, Poncet S, Taha MK, Larribe M, Darbon E, Khemiri A, Galinier A, Deutscher J. Boël G, et al. J Mol Microbiol Biotechnol. 2003;5(4):206-15. doi: 10.1159/000071072. J Mol Microbiol Biotechnol. 2003. PMID: 12867744 Review.
Cited by
- Pyrophosphate-producing protein dephosphorylation by HPr kinase/phosphorylase: a relic of early life?
Mijakovic I, Poncet S, Galinier A, Monedero V, Fieulaine S, Janin J, Nessler S, Marquez JA, Scheffzek K, Hasenbein S, Hengstenberg W, Deutscher J. Mijakovic I, et al. Proc Natl Acad Sci U S A. 2002 Oct 15;99(21):13442-7. doi: 10.1073/pnas.212410399. Epub 2002 Oct 1. Proc Natl Acad Sci U S A. 2002. PMID: 12359880 Free PMC article. - In vivo activity of enzymatic and regulatory components of the phosphoenolpyruvate:sugar phosphotransferase system in Mycoplasma pneumoniae.
Halbedel S, Hames C, Stülke J. Halbedel S, et al. J Bacteriol. 2004 Dec;186(23):7936-43. doi: 10.1128/JB.186.23.7936-7943.2004. J Bacteriol. 2004. PMID: 15547265 Free PMC article. - CcpA-dependent carbon catabolite repression in bacteria.
Warner JB, Lolkema JS. Warner JB, et al. Microbiol Mol Biol Rev. 2003 Dec;67(4):475-90. doi: 10.1128/MMBR.67.4.475-490.2003. Microbiol Mol Biol Rev. 2003. PMID: 14665673 Free PMC article. Review. - Transcriptional activator YesS is stimulated by histidine-phosphorylated HPr of the Bacillus subtilis phosphotransferase system.
Poncet S, Soret M, Mervelet P, Deutscher J, Noirot P. Poncet S, et al. J Biol Chem. 2009 Oct 9;284(41):28188-28197. doi: 10.1074/jbc.M109.046334. Epub 2009 Aug 3. J Biol Chem. 2009. PMID: 19651770 Free PMC article. - Pre- and post-docking sampling of conformational changes using ClustENM and HADDOCK for protein-protein and protein-DNA systems.
Kurkcuoglu Z, Bonvin AMJJ. Kurkcuoglu Z, et al. Proteins. 2020 Feb;88(2):292-306. doi: 10.1002/prot.25802. Epub 2019 Sep 3. Proteins. 2020. PMID: 31441121 Free PMC article.
References
- Audette G.F., Engelmann,R., Hengstenberg,W., Deutscher,J., Hayakawa,K., Quail,J.W. and Delbaere,L.T. (2000) The 1.9 Å resolution structure of phospho-serine 46 HPr from Enterococcus faecalis. J. Mol. Biol., 303, 545–553. - PubMed
- Berry M.B. and Phillips,G.N.,Jr (1998) Crystal structures of Bacillus stearothermophilus adenylate kinase with bound Ap5A, Mg2+ Ap5A and Mn2+ Ap5A reveal an intermediate lid position and six coordinate octahedral geometry for bound Mg2+ and Mn2+. Proteins, 32, 276–288. - PubMed
- Briozzo P., Golinelli-Pimpaneau,B., Gilles,A.M., Gaucher,J.F., Burlacu-Miron,S., Sakamoto,H., Janin,J. and Barzu,O. (1998) Structures of Escherichia coli CMP kinase alone and in complex with CDP: a new fold of the nucleoside monophosphate binding domain and insights into cytosine nucleotide specificity. Structure, 6, 1517–1527. - PubMed
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources