Roles for 147 embryonic lethal genes on C.elegans chromosome I identified by RNA interference and video microscopy - PubMed (original) (raw)
Roles for 147 embryonic lethal genes on C.elegans chromosome I identified by RNA interference and video microscopy
P Zipperlen et al. EMBO J. 2001.
Abstract
Early embryonic development involves complex events such as the regulation of cell division and the establishment of embryonic polarity. To identify genes involved in these events, we collected four-dimensional time-lapse video recordings of the first three cell divisions and analysed terminal phenotypes after RNA interference of 147 embryonic lethal genes previously identified in a systematic screen of Caenorhabditis elegans chromosome I. Over half gave defects in early processes such as meiosis, the assembly or position of the first mitotic spindle, cytokinesis, and proper nuclear positioning. For some phenotypic classes, the majority of genes are involved in a shared biochemical process. In addition, we identified loss-of-function phenotypes for genes of unknown function, but for which homologues exist in other organisms, shedding light on the function of these uncharacterized genes. When applied to the whole genome, this approach should identify the vast majority of genes required for early cell processes, paving the way for a greatly improved understanding of these processes and their regulation at the molecular level.
Figures
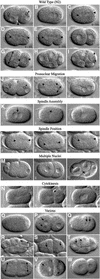
Fig. 1. Wild-type and RNAi mutant embryos. (A–I) Development of a wild-type embryo from the one- to the eight-cell stage. After fertilization, the oocyte nucleus completes meiosis and two polar bodies are extruded (not shown). (A) Subsequently the female and male pronuclei become visible. (A and B) The single oocyte pronucleus migrates to meet the sperm pronucleus in the posterior of the one cell embryo; this is accompanied by a pseudocleavage furrow [arrowhead in (A)]. (C–E) The first mitotic spindle is set up, elongating in the posterior direction through a rocking movement (arrowheads show ends of the spindle). (F) Two-cell embryo; first cleavage results in a larger anterior cell (AB) and a smaller posterior cell (P1). (G) The anterior AB cell divides before P1, along the short axis of the egg (AB spindle is shown by a double-headed arrow); P1 will divide along the anterior–posterior axis due to nucleocentrosomal rotation, which places centrosomes along this axis (arrowheads). (H) Four-cell embryo. (I) Division of P2 cell; the double-headed arrow shows orientation of the spindle. (J–N) Three time points from video recordings of each of five different RNAi mutants. (J) Pronuclear Migration; T06G6.9. Pronuclei fail to migrate. The embryo in (Jii) is similar stage to (C); pronuclear envelopes have broken down and sperm pronucleus has set up a small spindle (double-headed arrow). (K) Spindle Assembly; F32H2.3. (Kii) The mitotic spindle fails to set up [the arrowhead marks a small clear area where pronuclei have broken down; compare to spindle in (D)]. Note that the pronuclei meet centrally [arrow in (Ki)]. (L) Spindle Position; F21C3.5. The position of the mitotic spindle (marked by arrowheads) is unstable. Compare (Li–Liii) to (C–E). Individual images have been taken at 30 s intervals. (M) Multiple Nuclei; F56C11.5. Two cells each have two nuclei after division [arrows in (Miii)]. (N) Cytokinesis; Y18D10A.17. A normal cytokinesis furrow [arrow in (Nii)] regresses (Niii). (O–W) Single pictures illustrating other phenotypes. (O) 1-Cell Arrest; R12E2.3. (P) Meiosis; M01E11.6. Multiple female pronuclei are visible (arrows). (Q) Pronuclear Envelope; F56A3.3. The pronuclei have an indistinct appearance in comparison to (B). (R) Spindle Orientation; Y18D10A.5. Embryos have an abnormal P2 spindle orientation [compare to (I)]. (S) Nuclear Position; Y65B4B_10.b. Nuclei (arrows) are not centrally located in comparison with (F). (T) Nuclear Morphology; F33H2.5. Nuclei (arrows) are irregularly shaped and somewhat indistinct in comparison to the clear round nuclei in (H). (U) Spindle Orientation/Position; F20G4.3. AB and P1 cells divide at the same time, and both spindles exhibit a transverse orientation [compare to (G)]. The identical size of the two cells is reflective of a centrally located P0 spindle. (V) Aster Morphology (from Multiple Nuclei class); F26E4.1. Asters are very large and can easily be seen as clear regions next to the nuclei (marked by arrows). (W) Cytoplasmic Appearance; F26H9.6. The cytoplasm appears clear and less granular than wild type (F). Table I gives further information about the examples shown here. In all images, posterior is to the right.
Fig. 2. Distribution of genes among phenotypic and functional classes. (A) The first column gives phenotypic classes. EEP, early embryonic phenotype; LEP, late embryonic phenotype. Each row gives the number of genes with the given RNAi phenotype in each functional class, the total number in that class identified in this screen, and the total number in that class expected to be found in the genome by RNAi (see Materials and methods for the latter calculation). (B) Pie charts showing the distribution of functional classes among genes within the EEP, LEP <500 and LEP >500 phenotypic groupings. Functional classes were used as defined in Fraser et al. (2000).
Fig. 3. Terminal phenotype classes. (A) Wild-type embryo at 3-fold stage of elongation. The pharyngeal grinder is marked with an arrowhead. (B) <200 cell arrest; K02F2.3. (C) 200–500 cell arrest; F30F8.8. (D) Differentiated, not enclosed; T23D8.4. The embryo is not enclosed within epidermal tissue [compare with (E)] such that pharyngeal tissue (arrow) and some gut cells (arrowheads) are found at the surface of the embryo. (E) Enclosed; C32F10.5. Pharynx (arrow) is surrounded by epidermal tissue. (F) 1- to 2-fold arrest; F53B8.1. Pharynx is properly formed with a normal grinder (arrowhead), but the embryo is not properly elongated [compare with (A)].
Similar articles
- Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans.
Sönnichsen B, Koski LB, Walsh A, Marschall P, Neumann B, Brehm M, Alleaume AM, Artelt J, Bettencourt P, Cassin E, Hewitson M, Holz C, Khan M, Lazik S, Martin C, Nitzsche B, Ruer M, Stamford J, Winzi M, Heinkel R, Röder M, Finell J, Häntsch H, Jones SJ, Jones M, Piano F, Gunsalus KC, Oegema K, Gönczy P, Coulson A, Hyman AA, Echeverri CJ. Sönnichsen B, et al. Nature. 2005 Mar 24;434(7032):462-9. doi: 10.1038/nature03353. Nature. 2005. PMID: 15791247 - The Caenorhabditis elegans gene ncc-1 encodes a cdc2-related kinase required for M phase in meiotic and mitotic cell divisions, but not for S phase.
Boxem M, Srinivasan DG, van den Heuvel S. Boxem M, et al. Development. 1999 May;126(10):2227-39. doi: 10.1242/dev.126.10.2227. Development. 1999. PMID: 10207147 - A genome-wide RNAi screen for enhancers of par mutants reveals new contributors to early embryonic polarity in Caenorhabditis elegans.
Morton DG, Hoose WA, Kemphues KJ. Morton DG, et al. Genetics. 2012 Nov;192(3):929-42. doi: 10.1534/genetics.112.143727. Epub 2012 Aug 10. Genetics. 2012. PMID: 22887819 Free PMC article. - High-throughput reverse genetics: RNAi screens in Caenorhabditis elegans.
Bargmann CI. Bargmann CI. Genome Biol. 2001;2(2):REVIEWS1005. doi: 10.1186/gb-2001-2-2-reviews1005. Epub 2001 Jan 31. Genome Biol. 2001. PMID: 11182891 Free PMC article. Review. - Specification and development of the germline in Caenorhabditis elegans.
Strome S, Garvin C, Paulsen J, Capowski E, Martin P, Beanan M. Strome S, et al. Ciba Found Symp. 1994;182:31-45; discussion 45-57. doi: 10.1002/9780470514573.ch3. Ciba Found Symp. 1994. PMID: 7835156 Review.
Cited by
- Anaphase-promoting complex in Caenorhabditis elegans.
Yeong FM. Yeong FM. Mol Cell Biol. 2004 Mar;24(6):2215-25. doi: 10.1128/MCB.24.6.2215-2225.2004. Mol Cell Biol. 2004. PMID: 14993261 Free PMC article. Review. No abstract available. - Identification of the C. elegans anaphase promoting complex subunit Cdc26 by phenotypic profiling and functional rescue in yeast.
Dong Y, Bogdanova A, Habermann B, Zachariae W, Ahringer J. Dong Y, et al. BMC Dev Biol. 2007 Mar 20;7:19. doi: 10.1186/1471-213X-7-19. BMC Dev Biol. 2007. PMID: 17374146 Free PMC article. - Ran GTPase cycle and importins alpha and beta are essential for spindle formation and nuclear envelope assembly in living Caenorhabditis elegans embryos.
Askjaer P, Galy V, Hannak E, Mattaj IW. Askjaer P, et al. Mol Biol Cell. 2002 Dec;13(12):4355-70. doi: 10.1091/mbc.e02-06-0346. Mol Biol Cell. 2002. PMID: 12475958 Free PMC article. - Peroxisome protein transportation affects metabolism of branched-chain fatty acids that critically impact growth and development of C. elegans.
Wang R, Kniazeva M, Han M. Wang R, et al. PLoS One. 2013 Sep 27;8(9):e76270. doi: 10.1371/journal.pone.0076270. eCollection 2013. PLoS One. 2013. PMID: 24086720 Free PMC article. - Generation of an external guide sequence library for a reverse genetic screen in Caenorhabditis elegans.
Yan Q, Zhao R, Zheng W, Yin C, Zhang B, Ma W. Yan Q, et al. BMC Biotechnol. 2009 May 20;9:47. doi: 10.1186/1472-6750-9-47. BMC Biotechnol. 2009. PMID: 19457250 Free PMC article.
References
- Draper B.W., Mello,C.C., Bowerman,B., Hardin,J. and Priess,J.R. (1996) MEX-3 is a KH domain protein that regulates blastomere identity in early C. elegans embryos. Cell, 87, 205–216. - PubMed
- Edgar L.G., Wolf,N. and Wood,W.B. (1994) Early transcription in Caenorhabditis elegans embryos. Development, 120, 443–451. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources