Flexible use of high-density oligonucleotide arrays for single-nucleotide polymorphism discovery and validation - PubMed (original) (raw)
Comparative Study
. 2001 Aug;11(8):1418-24.
doi: 10.1101/gr.171101.
Affiliations
- PMID: 11483583
- PMCID: PMC311102
- DOI: 10.1101/gr.171101
Comparative Study
Flexible use of high-density oligonucleotide arrays for single-nucleotide polymorphism discovery and validation
S Dong et al. Genome Res. 2001 Aug.
Abstract
A method for identifying and validating single nucleotide polymorphisms (SNPs) with high-density oligonucleotide arrays without the need for locus-specific polymerase chain reactions (PCR) is described in this report. Genomic DNAs were divided into subsets with complexity of ~10 Mb by restriction enzyme digestion and gel-based fragment size resolution, ligated to a common adaptor, and amplified with one primer in a single PCR reaction. As a demonstration of this approach, a total of 124 SNPs were located in 190 kb of genomic sequences distributed across the entire human genome by hybridizing to high-density variant detection arrays (VDA). A set of independent validation experiments was conducted for these SNPs employing bead-based affinity selection followed by hybridization of the affinity-selected SNP-containing fragments to the same VDA that was used to identify the SNPs. A total of 98.7% (74/75) of these SNPs were confirmed using both DNA dideoxynucleotide sequencing and the VDA methodologies. With flexible sample preparation, high-density oligonucleotide arrays can be tailored for even larger scale genome-wide SNP discovery as well as validation.
Figures
Figure 1
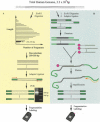
Schemes of (A) single nucleotide polymorphism (SNP) discovery and (B) validation. (A) Genomic DNA was digested with _Eco_RI and separated by size on agarose gel. A 250–350 bp fraction was cut from the gel, extracted, and ligated to an adaptor. The ligated DNA was amplified with one primer that interrogated the adaptor sequence. SNPs were screened by hybridizins to VDA. (B) Designated targets were enriched from total genomic DNA by oligonucleotides bound to magnetic beads that interrogated the SNP sites. The genomic DNA was digested with _Eco_RI and ligated with an adaptor. The enriched targets were amplified with the same primer and screened similarly as in A.
Figure 2
SNP screening with variant detection arrays (VDAs). (A) The whole image of 40 pM targets hybridized with VDA. (B) An expanded view of a portion of A. (C) The same region of image as in B except that the chip was hybridized with enriched targets. (D) A single nucleotide polymorphism (SNP) detected and confirmed by alternative sample preparation. From top to bottom, the wild type (base T at the center), heterozygous SNP (bases C and T at the center), and homozygous mutation (base C at the center) are shown.
Figure 3
Suppression of signals of repetitve sequences with human Cot-1 fraction. The variant detection arrays (VDAs) were hybridized with enriched targets without adding Cot-1 (A) and with 30 ng/μL human Cot-1 fraction (B). The bright signals at the top of the image are attributable to unique sequences that were enriched. The dark region below had little intensity because the sequences were not enriched. The lower portion shows the cross-hybridization from repetitive sequences (A) that was diminished by adding human Cot-1 DNA.
Similar articles
- Large-scale genotyping of complex DNA.
Kennedy GC, Matsuzaki H, Dong S, Liu WM, Huang J, Liu G, Su X, Cao M, Chen W, Zhang J, Liu W, Yang G, Di X, Ryder T, He Z, Surti U, Phillips MS, Boyce-Jacino MT, Fodor SP, Jones KW. Kennedy GC, et al. Nat Biotechnol. 2003 Oct;21(10):1233-7. doi: 10.1038/nbt869. Epub 2003 Sep 7. Nat Biotechnol. 2003. PMID: 12960966 - Direct selection of human genomic loci by microarray hybridization.
Albert TJ, Molla MN, Muzny DM, Nazareth L, Wheeler D, Song X, Richmond TA, Middle CM, Rodesch MJ, Packard CJ, Weinstock GM, Gibbs RA. Albert TJ, et al. Nat Methods. 2007 Nov;4(11):903-5. doi: 10.1038/nmeth1111. Epub 2007 Oct 14. Nat Methods. 2007. PMID: 17934467 - Combined array-comparative genomic hybridization and single-nucleotide polymorphism-loss of heterozygosity analysis reveals complex genetic alterations in cervical cancer.
Kloth JN, Oosting J, van Wezel T, Szuhai K, Knijnenburg J, Gorter A, Kenter GG, Fleuren GJ, Jordanova ES. Kloth JN, et al. BMC Genomics. 2007 Feb 20;8:53. doi: 10.1186/1471-2164-8-53. BMC Genomics. 2007. PMID: 17311676 Free PMC article. - Analysis of SNPs and other genomic variations using gel-based chips.
Kolchinsky A, Mirzabekov A. Kolchinsky A, et al. Hum Mutat. 2002 Apr;19(4):343-60. doi: 10.1002/humu.10077. Hum Mutat. 2002. PMID: 11933189 Review. - [Advances in high-density whole genome-wide single nucleotide polymorphism array in cancer research].
Zeng ZY, Xiong W, Zhou YH, Li XL, Li GY. Zeng ZY, et al. Ai Zheng. 2006 Nov;25(11):1454-8. Ai Zheng. 2006. PMID: 17094921 Review. Chinese.
Cited by
- Parallel genotyping of over 10,000 SNPs using a one-primer assay on a high-density oligonucleotide array.
Matsuzaki H, Loi H, Dong S, Tsai YY, Fang J, Law J, Di X, Liu WM, Yang G, Liu G, Huang J, Kennedy GC, Ryder TB, Marcus GA, Walsh PS, Shriver MD, Puck JM, Jones KW, Mei R. Matsuzaki H, et al. Genome Res. 2004 Mar;14(3):414-25. doi: 10.1101/gr.2014904. Genome Res. 2004. PMID: 14993208 Free PMC article. - Multiplex detection and genotyping of point mutations involved in charcot-marie-tooth disease using a hairpin microarray-based assay.
Baaj Y, Magdelaine C, Ubertelli V, Valat C, Mousseau Y, Qiu H, Funalot B, Vallat JM, Sturtz FG. Baaj Y, et al. Res Lett Biochem. 2009;2009:960560. doi: 10.1155/2009/960560. Epub 2009 Jun 11. Res Lett Biochem. 2009. PMID: 22820753 Free PMC article. - SNPs by AFLP (SBA): a rapid SNP isolation strategy for non-model organisms.
Nicod JC, Largiadèr CR. Nicod JC, et al. Nucleic Acids Res. 2003 Mar 1;31(5):e19. doi: 10.1093/nar/gng019. Nucleic Acids Res. 2003. PMID: 12595568 Free PMC article. - Validation and extension of an empirical Bayes method for SNP calling on Affymetrix microarrays.
Lin S, Carvalho B, Cutler DJ, Arking DE, Chakravarti A, Irizarry RA. Lin S, et al. Genome Biol. 2008 Apr 3;9(4):R63. doi: 10.1186/gb-2008-9-4-r63. Genome Biol. 2008. PMID: 18387188 Free PMC article. - Single-feature polymorphism discovery in the barley transcriptome.
Rostoks N, Borevitz JO, Hedley PE, Russell J, Mudie S, Morris J, Cardle L, Marshall DF, Waugh R. Rostoks N, et al. Genome Biol. 2005;6(6):R54. doi: 10.1186/gb-2005-6-6-r54. Epub 2005 May 11. Genome Biol. 2005. PMID: 15960806 Free PMC article.
References
- Cargill M, Altshuler D, Ireland J, Sklar P, Ardlie K, Patil N, Lane CR, Lim EP, Kalyanaraman N, Nemesh J, et al. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat Genet. 1999;22:231–238. - PubMed
- Chee M, Yang R, Hubbell E, Berno A, Huang XC, Stern D, Winkler J, Lockhart DJ, Morris MS, Fodor SPS. Accessing genetic information with high-density DNA arrays. Science. 1996;274:610–614. - PubMed
- Collins FS, Guyer MS, Charkravarti A. Variations in a theme: Cataloging human DNA sequence variation. Science. 1997;278:1580–1581. - PubMed
- Cooper DN, Smith BA, Cooke HJ, Niemann S, Schmidtke J. An estimate of unique DNA sequence heterozygosity in the human genomes. Hum Genet. 1985;69:201–205. - PubMed
- Hacia JG, Fan J-B, Ryder O, Jin L, Edgemon K, Ghandour G, Mayer RA, Sun B, Hsie L, Robins CM, et al. Determination of ancestral alleles for human single-nucleotide polymorphisms using high-density oligonucleotide arrays. Nat Genet. 1999;22:164–167. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous