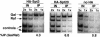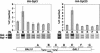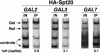The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4 - PubMed (original) (raw)
The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4
E Larschan et al. Genes Dev. 2001.
Abstract
Previous studies demonstrated that the SAGA (Spt-Ada-Gcn5-Acetyltransferase) complex facilitates the binding of TATA-binding protein (TBP) during transcriptional activation of the GAL1 gene of Saccharomyces cerevisiae. TBP binding was shown to require the SAGA components Spt3 and Spt20/Ada5, but not the SAGA component Gcn5. We have now examined whether SAGA is directly required as a coactivator in vivo by using chromatin immunoprecipitation analysis. Our results demonstrate that SAGA is physically recruited in vivo to the upstream activation sequence (UAS) regions of the galactose-inducible GAL genes. This recruitment is dependent on both induction by galactose and the Gal4 activation domain. Furthermore, we demonstrate that another well-characterized activator, Gal4-VP16, also recruits SAGA in vivo. Finally, we provide evidence that a specific interaction between Spt3 and TBP in vivo is important for Gal4 transcriptional activation at a step after SAGA recruitment. These results, taken together with previous studies, demonstrate a dependent pathway for the recruitment of TBP to GAL gene promoters consisting of the recruitment of SAGA by Gal4 and the subsequent recruitment of TBP by SAGA.
Figures
Figure 1
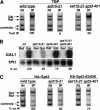
Spt3 and Spt20 are recruited to the GAL1–GAL10 UASG in the presence of galactose. Chromatin immunoprecipitation was performed on wild-type strains containing HA1–Spt20 (FY1977), HA1–Spt3 (FY1978), or an untagged isogenic strain as a negative control (FY1976). Chromatin immunoprecipitation with the 12CA5 anti-HA antibody was conducted on each strain grown both in raffinose (Raf) and after a 20-min galactose induction (Gal; see Materials and Methods). Quantitation was conducted as described in Materials and Methods. The following % IP (Gal/Raf) values are reported with standard error: HA1–Spt3 (4.3 ± 1.2, six experiments); HA1–Spt20 (6.8 ± 1.2, four experiments); No HA1 (0.8 ± 0.1, three experiments). The GAL1–GAL10 UASG PCR product spans from −536 to −276 relative to the +1 of translation for GAL1.
Figure 2
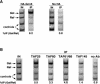
Gcn5 and the SAGA TATA-binding-protein associated factors (TAFs) are bound to the UASG in a galactose-dependent manner. (A) Gcn5 is recruited to the GAL1–GAL10 UASG in a galactose-dependent manner. Chromatin immunoprecipitation was conducted on a strain containing HA1–Gcn5 (FY1980) and an untagged isogenic strain (FY1981) by using the 12CA5 anti-HA antibody. The GAL1–10 UASG PCR primers were used (Fig. 1). The percent immunoprecipitated (IP) (Gal/Raf) values are averages derived from three independent chromatin immunoprecipitation experiments: HA1–Gcn5 (9.2 +/- 0.9); No HA1 (0.8 ± 0.2). (B) SAGA TAFs are recruited to the GAL1–10 UASG, whereas a TFIID-specific TAF is not recruited. Chromatin immunoprecipitation analysis was conducted on a wild-type strain (FY1978) by using antibodies against TAF25, TAF60, TAF61/68, and TAF145 previously described by Li et al. (2000). The GAL1–10 UASG PCR primers were used (Fig. 1). Percent IP (Gal/Raf) values are averages derived from three independent chromatin extracts: TAF25 (5.5 ± 1.0); TAF60 (3.3 ± 0.2); TAF61/68 (4.8 ± 0.8); TAF145 (1.4 ± 0.6); no Ab (0.9 ± 0.2).
Figure 3
Spt3 and Spt20 localize specifically to the GAL1–GAL10 UASG. PCR products A–G span the GAL10 5′, GAL1–GAL10 UASG, GAL1 ORF, and GAL1 3′ UTR. The locations of the PCR products relative to the +1 of translation are as follows: GAL10: A (−58 to +182); GAL1: B (−536 to −276); C (−190 to +54); D (+271 to +596); E (+590 to +877); F (+980 to +1309); G (+1370 to +1657). The translation termination codon for GAL1 is at +1588. Inputs shown are representative of those obtained for PCR products A–G and control PCR products are not shown.
Figure 4
SAGA is recruited to GAL2, GAL3, and GAL7 UAS sequences. Chromatin immunoprecipitation was conducted on a strain containing HA1–Spt20 (FY1977) by using the 12CA5 anti-HA antibody as described for Figure 1. PCR products span the GAL2, GAL3, and GAL7 UAS regions as follows: GAL2 UAS (−464 to −195); GAL3 UAS (−420 to −157); and GAL7 UAS (−296 to −54). Percent IP (Gal/Raf) values are averages derived from two chromatin extracts: GAL2 UAS (5.8 ± 1.4); GAL3 UAS (3.1 ± 0.6); GAL7 UAS (6.7 ± 0.7). The lower level of the GAL7 PCR product compared with the controls is probably caused by inefficient PCR with that pair of primers.
Figure 5
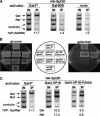
The VP16 activation domain recruits SAGA to the GAL1–10 UASG. (A) The Gal4 activation domain is required for SAGA recruitment to the GAL1–10 UAS. Chromatin immunoprecipitation experiments were conducted on the following strains by using the 12CA5 anti-HA antibody: 1) Gal4+: HA1–Spt20 + Vector (FY1977 + pRS415); 2) Gal4DB: HA1–Spt20 _gal4_Δ + GAL4DB ( FY1982 + pPC97); 3) none: HA1–Spt20 _gal4_Δ + Vector (FY1982 + pRS415). Strains were grown in SC-Leu Raf media and then induced with galactose for 20 min before cross-linking. The GAL1–GAL10 UASG PCR primers were used (Fig. 1). Percent IP (Gal/Raf) values with standard error are as follows: Gal4+ (11.7 ± 0.4); Gal4DB (1.2 ± 0.1); none (1.0 ± 0.1). (B) Gal4–VP16 requires SAGA for activation. Strains containing the Gal4+, Gal4–VP16, or Gal4–VP16–F442A activators were tested for their ability to grow on glucose and galactose as carbon sources and for their dependence on Spt20 for activation. Strains were tested for growth on SC-Leu plates containing glucose or galactose by replica plating and are shown after 4 d of growth at 30°C. The SPT20 allele and the activator present in each strain is shown in the figure. (C) Gal4–VP16 recruits SAGA to the _GAL1_-GAL10 UAS. Chromatin immunoprecipitation experiments were conducted as described for Fig. 5A. Spt20 recruitment was assayed in a wild-type strain and in an HA1–Spt20 _gal4_Δ strain (FY1982) also containing either Gal4–VP16 or Gal4–VP16–F442A plasmids. Quantitation was performed as described in Materials and Methods. Individual values for % IP (Gal/control) and % IP (Raf/control) are reported because both wild-type and mutant VP16 activation domains activate under galactose and raffinose growth conditions. The following values include standard error: Gal4 (Gal/Raf :11.7 ± 0.4); Gal4–VP16 (Gal: 10.5 ± 1.5; Raf: 17.3 ± 1.6); Gal4–VP16–F442A (Gal: 3.0 ± 1.2; Raf: 6.8 ± 2.9).
Figure 6
Spt3 is not required for SAGA recruitment. Chromatin immunoprecipitation was conducted by using the anti-HA antibody 12CA5 on a strain containing HA1–Spt20 _spt3_Δ (FY1984) and the wild-type HA1–Spt20 strain (FY1977) as a positive control. Quantitation was conducted as described in Materials and Methods and the average % IP value for the _spt3_Δ mutant was normalized to the value for the wild-type strain. The average % IP (Gal) values for the wild-type and _spt3_Δ mutant strains were calculated from three independent experiments.
Figure 7
An Spt3–TATA binding protein (TBP) functional interaction facilitates TBP binding to the GAL1 TATA box. (A) Chromatin immunoprecipitation by using the anti-TBP antibody was conducted on the following strains: HA1–Spt3 wild-type (FY1978), HA1–Spt3 spt15-21 (FY1985), and HA1–spt3-401 spt15-21 (FY1987). Binding to the GAL1 TATA box was assayed by quantitating a PCR product spanning GAL1 TATA (Fig. 3; PCR Product C). Calculations were conducted as described for Fig. 6. Average % IP (Gal) values for wild-type and mutant strains were calculated from four independent experiments. (B) Northern analysis of GAL1 mRNA levels in wild-type, spt15-21, spt3-401, and spt3-401 spt15-21 strains. Northern blot analyses were conducted on the same cultures used for chromatin immunoprecipitations shown in Fig. 7A. Quantitation was done with the PhosphorImager (Molecular Dynamics). Values obtained for the intensity of each GAL1 band were normalized to the TPI1 probe loading control signal for the same sample. Then the percent of GAL1 mRNA, normalized to the wild-type value, was determined for each mutant. The averages from four independent experiments are reported for each strain: spt15-21 (25 ± 7); spt3-401 (74 ± 5); spt15-21 spt3-401 (80 ± 13). (C) Spt3 recruitment to the GAL1–10 UAS is unaffected in the spt15-21 mutant. Chromatin immunoprecipitation was conducted as described for Fig. 7A except the 12CA5 anti-HA antibody was used for chromatin immunoprecipitation. Calculations were performed as described for Fig. 6. Average % IP (Gal) values for wild-type and mutant strains were calculated from four independent experiments.
Figure 8
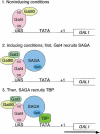
A model for SAGA functioning as a coactivator for Gal4 by facilitating TATA-binding-protein (TBP) binding to the TATA box of the GAL1 gene. (1) Under noninducing conditions, Gal4 is bound to the UASG via its DNA-binding domain (DB), and the Gal4 activation domain (AD) is blocked by Gal80. (2) After the addition of galactose, the Gal3 inducer is activated and alters the Gal80–Gal4 complex such that the Gal4 activation domain is no longer blocked by Gal80. This change allows the Gal4 activation domain to recruit SAGA to UASG. The presence of the Gal3 protein at the promoter is suggested by the formation of a Gal3–Gal4–Gal80 complex in vitro and in vivo (Chasman and Kornberg 1990; Leuther and Johnston 1992; Parthun and Jaehning 1992; Platt and Reece 1998; Sil et al. 1999). However, other data have suggested that Gal3 is cytoplasmically localized (Peng and Hopper 2000). (3) Once recruited to the promoter, SAGA, mainly via an Spt3-–TBP interaction, recruits TBP to the TATA box to allow transcription initiation.
Similar articles
- Differential requirement of SAGA components for recruitment of TATA-box-binding protein to promoters in vivo.
Bhaumik SR, Green MR. Bhaumik SR, et al. Mol Cell Biol. 2002 Nov;22(21):7365-71. doi: 10.1128/MCB.22.21.7365-7371.2002. Mol Cell Biol. 2002. PMID: 12370284 Free PMC article. - SAGA is an essential in vivo target of the yeast acidic activator Gal4p.
Bhaumik SR, Green MR. Bhaumik SR, et al. Genes Dev. 2001 Aug 1;15(15):1935-45. doi: 10.1101/gad.911401. Genes Dev. 2001. PMID: 11485988 Free PMC article. - The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo.
Dudley AM, Rougeulle C, Winston F. Dudley AM, et al. Genes Dev. 1999 Nov 15;13(22):2940-5. doi: 10.1101/gad.13.22.2940. Genes Dev. 1999. PMID: 10580001 Free PMC article. - Recruitment of chromatin remodelling factors during gene activation via the glucocorticoid receptor N-terminal domain.
Wallberg AE, Flinn EM, Gustafsson JA, Wright AP. Wallberg AE, et al. Biochem Soc Trans. 2000;28(4):410-4. Biochem Soc Trans. 2000. PMID: 10961930 Review. - A SAGA of histone acetylation and gene expression.
Hampsey M. Hampsey M. Trends Genet. 1997 Nov;13(11):427-9. doi: 10.1016/s0168-9525(97)01292-4. Trends Genet. 1997. PMID: 9385836 Review. No abstract available.
Cited by
- Regulation of TATA-binding protein binding by the SAGA complex and the Nhp6 high-mobility group protein.
Yu Y, Eriksson P, Bhoite LT, Stillman DJ. Yu Y, et al. Mol Cell Biol. 2003 Mar;23(6):1910-21. doi: 10.1128/MCB.23.6.1910-1921.2003. Mol Cell Biol. 2003. PMID: 12612066 Free PMC article. - Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry.
Sanders SL, Jennings J, Canutescu A, Link AJ, Weil PA. Sanders SL, et al. Mol Cell Biol. 2002 Jul;22(13):4723-38. doi: 10.1128/MCB.22.13.4723-4738.2002. Mol Cell Biol. 2002. PMID: 12052880 Free PMC article. - STAGA recruits Mediator to the MYC oncoprotein to stimulate transcription and cell proliferation.
Liu X, Vorontchikhina M, Wang YL, Faiola F, Martinez E. Liu X, et al. Mol Cell Biol. 2008 Jan;28(1):108-21. doi: 10.1128/MCB.01402-07. Epub 2007 Oct 29. Mol Cell Biol. 2008. PMID: 17967894 Free PMC article. - SALSA, a variant of yeast SAGA, contains truncated Spt7, which correlates with activated transcription.
Sterner DE, Belotserkovskaya R, Berger SL. Sterner DE, et al. Proc Natl Acad Sci U S A. 2002 Sep 3;99(18):11622-7. doi: 10.1073/pnas.182021199. Epub 2002 Aug 19. Proc Natl Acad Sci U S A. 2002. PMID: 12186975 Free PMC article. - Identification and characterization of Elf1, a conserved transcription elongation factor in Saccharomyces cerevisiae.
Prather D, Krogan NJ, Emili A, Greenblatt JF, Winston F. Prather D, et al. Mol Cell Biol. 2005 Nov;25(22):10122-35. doi: 10.1128/MCB.25.22.10122-10135.2005. Mol Cell Biol. 2005. PMID: 16260625 Free PMC article.
References
- Barlev NA, Candau R, Wang L, Darpino P, Silverman N, Berger SL. Characterization of physical interactions of the putative transcriptional adaptor, ADA2, with acidic activation domains and TATA-binding protein. J Biol Chem. 1995;270:19337–19344. - PubMed
- Berger SL, Pina B, Silverman N, Marcus GA, Agapite J, Regier JL, Triezenberg SJ, Guarente L. Genetic isolation of ADA2: A potential transcriptional adaptor required for function of certain acidic activation domains. Cell. 1992;70:251–265. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials