Inhibition of acetyl coenzyme A carboxylase activity restores expression of the INO1 gene in a snf1 mutant strain of Saccharomyces cerevisiae - PubMed (original) (raw)
Inhibition of acetyl coenzyme A carboxylase activity restores expression of the INO1 gene in a snf1 mutant strain of Saccharomyces cerevisiae
M K Shirra et al. Mol Cell Biol. 2001 Sep.
Abstract
Mutations in the Saccharomyces cerevisiae SNF1 gene affect a number of cellular processes, including the expression of genes involved in carbon source utilization and phospholipid biosynthesis. To identify targets of the Snf1 kinase that modulate expression of INO1, a gene required for an early, rate-limiting step in phospholipid biosynthesis, we performed a genetic selection for suppressors of the inositol auxotrophy of snf1Delta strains. We identified mutations in ACC1 and FAS1, two genes important for fatty acid biosynthesis in yeast; ACC1 encodes acetyl coenzyme A carboxylase (Acc1), and FAS1 encodes the beta subunit of fatty acid synthase. Acc1 was shown previously to be phosphorylated and inactivated by Snf1. Here we show that snf1Delta strains with increased Acc1 activity exhibit decreased INO1 transcription. Strains carrying the ACC1 suppressor mutation have reduced Acc1 activity in vitro and in vivo, as revealed by enzymatic assays and increased sensitivity to the Acc1-specific inhibitor soraphen A. Moreover, a reduction in Acc1 activity, caused by addition of soraphen A, provision of exogenous fatty acid, or conditional expression of ACC1, suppresses the inositol auxotrophy of snf1Delta strains. Together, these findings indicate that the inositol auxotrophy of snf1Delta strains arises in part from elevated Acc1 activity and that a reduction in this activity restores INO1 expression in these strains. These results reveal a Snf1-dependent connection between fatty acid production and phospholipid biosynthesis, identify Acc1 as a Snf1 target important for INO1 transcription, and suggest models in which metabolites that are generated or utilized during fatty acid biosynthesis can significantly influence gene expression in yeast.
Figures
FIG. 1
Suppressor mutations significantly increase transcription of INO1 in strains containing snf1_Δ_10. Northern analysis of INO1 transcription is shown. Repressed RNA samples (R) were obtained from cells grown in −I media supplemented with 200 μM inositol. Derepressed RNA samples (DR) were obtained from cells that were grown in 200 μM inositol media, washed, resuspended in −I media supplemented with 10 μM inositol, and harvested after incubation at 30°C for an additional 10 h. Strains used were as follows: PY165 (lanes 1 and 2), PY133 (lanes 3 and 4), PY794 (lanes 5 and 6), PY802 (lanes 7 and 8), and PY803 (lanes 9 and 10). The filter from the upper panel was reprobed for SPT15 mRNA as a control. A representative experiment is shown.
FIG. 2
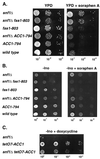
Suppression of the snf1_Δ_10 Ino− phenotype by ACC1 mutations is allele specific. Strains grown overnight in inositol-containing medium were harvested, washed, normalized by OD600, and spotted onto plates in a series of three 10-fold dilutions. All media contained 1% Brij 58 and either contained (+Ino) or lacked (−Ino) 75 μM inositol. Plates were allowed to grow for 3 days at 30°C. Strains used were as follows: YAXU008-3a, YAXU008-3d, YAXU008-3b, and YAXU009-6a.
FIG. 3
The effect of soraphen A and reduced ACC1 expression on the growth and inositol auxotrophy of snf1_Δ_10 strains. Yeast cultures, grown overnight in YPD, were diluted in sterile water to the OD600 indicated at the bottom of each lane, and 5-μl samples were spotted onto the following media: YPD and YPD plus 0.25 μg of soraphen A/ml (A); -Ino and -Ino plus 0.25 μg of soraphen A/ml (B); and -Ino + 2 μg of doxycycline/ml (C). The following yeast strains were tested: PY133, PY803, PY170, PY794, PY199, PY165, AUY009, and YAXU015-1a.
FIG. 4
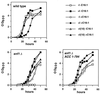
Palmitoleic acid suppresses the inositol auxotrophy of snf1_Δ_10 strains. Strains grown overnight in inositol-containing medium (+I −C16:1) were harvested, washed, and used to start liquid cultures at an OD600 of 0.01 for growth at 30°C. All media contained 1% Brij 58 and the indicated combinations of inositol (+I, 75 μM; −I, 0 μM) and palmitoleic acid (+C16:1, 0.5 mM; −C16:1, 0 mM). In the case of the snf1_Δ_10 mutant, the media contained 10 μM inositol instead of 0 μM inositol. Strains used were as follows: SNF1 (PY165), snf1_Δ_10 (PY133), fas1-803 (PY170), ACC1-794 (PY199), snf1_Δ_10 fas1-803 (PY803), and snf1_Δ_10 ACC1-794 (PY794).
FIG. 5
Fatty acid supplementation supports a high level of INO1 transcription in snf1_Δ_10 strains. Northern analysis of INO1 transcription is shown. Cells were grown in media containing 1% Brij 58 detergent in the presence or absence of 0.5 mM palmitoleic acid (C16:1) and the indicated concentrations of inositol. Cells were harvested at a cell density of 1 × 107 to 2 × 107 cells/ml. Strains used were as follows: PY165 (lanes 1 to 5), PY133 (lanes 6 to 9), PY794 (lanes 10 to 11), and PY199 (lanes 12 to 16). The filter from the upper panel was reprobed for TUB2 mRNA as a control. A representative experiment is shown.
FIG. 6
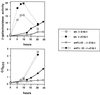
Effect of palmitoleic acid on the kinetics of INO1 derepression. Strains bearing the plasmid pJH359 (INO1-CYC1-lacZ) (47) were grown to mid-logarithmic phase in synthetic medium containing 1% Brij 58 and 75 μM inositol. Following harvesting and washing, each strain was used to inoculate two different media (−I −C16:1 and −I +C16:1) at an OD600 of ≈0.2. At various times, aliquots of the cultures were removed and assayed for β-galactosidase activity. Data represent the averages of results of two independent experiments. Strains used were as follows: SNF1 (PY165), snf1_Δ_10 (PY133), and snf1_Δ_10 ACC1-794 (PY794). The apparent decrease in β-galactosidase activity [_A_420 × 1,000/(min × ml × OD600)] in the wild-type culture at the 20-h time point is a reflection of the strain's continued growth, once its β-galactosidase activity has reached a plateau level.
FIG. 7
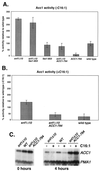
Acc1 enzyme activity and ACC1 expression in the absence and presence of exogenous palmitoleic acid. (A and B) Acc1 enzyme activity was determined as described in Materials and Methods. The activity was determined three to four times and normalized to the protein concentration in the homogenate, and it is depicted as specific activity relative to activity of a wild-type strain grown in the absence of exogenous C16:1 (set at 100%). For the experiment depicted in panel B, C16:1 was added to the growth media to a final concentration of 100 μM without detergent (detergent was found to interfere with the enzyme preparation and resulted in a loss of Acc1 activity). (C) Northern analysis of ACC1 expression. Total RNA was prepared 0 and 4 h after addition of C16:1 (100 μM, where indicated), separated on denaturing agarose gels, blotted, and hybridized with digoxigenin-labeled ACC1 and PMA1 probes. Strains used were as follows: PY133, PY803, PY170, PY794, PY199, and PY165.
FIG. 8
Schematic diagram of phospholipid biosynthesis in S. cerevisiae. Solid arrows indicate direct enzymatic conversions. Dashed arrows indicate conversions that require more than one enzymatic step. Gene designations are in bold italics. Phosphorylation of Acc1 by the SNF1 gene product inhibits Acc1 activity. Acyl-CoAs, including malonyl-CoA, palmitoyl-CoA, palmitoleoyl-CoA, stearoyl-CoA, and oleoyl-CoA, inhibit Acc1 activity. Externally added palmitate (palmitateext) and palmitoleate (palmitoleateext) are converted to their respective CoA derivatives in the cell. Lyso-PtdOH, lysophosphatidic acid; Gro-3-P, glycerol-3-phosphate; Gluc-6-P, glucose-6-phosphate; Ins-1-P, inositol-1-phosphate; PtdOH, phosphatidic acid; DAG, diacylglycerol; TAG, triacylglycerol; Etn, ethanolamine; Cho, choline; PtdIns, phosphatidylinositol; PtdSer, phosphatidylserine; PtdEtn, phosphatidylethanolamine; PtdCho, phosphatidylcholine; PIP's, polyphosphoinositides.
Similar articles
- Evidence for the involvement of the Glc7-Reg1 phosphatase and the Snf1-Snf4 kinase in the regulation of INO1 transcription in Saccharomyces cerevisiae.
Shirra MK, Arndt KM. Shirra MK, et al. Genetics. 1999 May;152(1):73-87. doi: 10.1093/genetics/152.1.73. Genetics. 1999. PMID: 10224244 Free PMC article. - The Snf1 protein kinase and Sit4 protein phosphatase have opposing functions in regulating TATA-binding protein association with the Saccharomyces cerevisiae INO1 promoter.
Shirra MK, Rogers SE, Alexander DE, Arndt KM. Shirra MK, et al. Genetics. 2005 Apr;169(4):1957-72. doi: 10.1534/genetics.104.038075. Epub 2005 Feb 16. Genetics. 2005. PMID: 15716495 Free PMC article. - SNF1/AMPK pathways in yeast.
Hedbacker K, Carlson M. Hedbacker K, et al. Front Biosci. 2008 Jan 1;13:2408-20. doi: 10.2741/2854. Front Biosci. 2008. PMID: 17981722 Free PMC article. Review. - Transcriptional regulation of yeast phospholipid biosynthetic genes.
Chen M, Hancock LC, Lopes JM. Chen M, et al. Biochim Biophys Acta. 2007 Mar;1771(3):310-21. doi: 10.1016/j.bbalip.2006.05.017. Epub 2006 Jun 6. Biochim Biophys Acta. 2007. PMID: 16854618 Review.
Cited by
- Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae.
Henry SA, Kohlwein SD, Carman GM. Henry SA, et al. Genetics. 2012 Feb;190(2):317-49. doi: 10.1534/genetics.111.130286. Genetics. 2012. PMID: 22345606 Free PMC article. Review. - The Effect of Lithium on the Budding Yeast Saccharomyces cerevisiae upon Stress Adaptation.
Reith P, Braam S, Welkenhuysen N, Lecinski S, Shepherd J, MacDonald C, Leake MC, Hohmann S, Shashkova S, Cvijovic M. Reith P, et al. Microorganisms. 2022 Mar 9;10(3):590. doi: 10.3390/microorganisms10030590. Microorganisms. 2022. PMID: 35336166 Free PMC article. - Gene recruitment of the activated INO1 locus to the nuclear membrane.
Brickner JH, Walter P. Brickner JH, et al. PLoS Biol. 2004 Nov;2(11):e342. doi: 10.1371/journal.pbio.0020342. Epub 2004 Sep 28. PLoS Biol. 2004. PMID: 15455074 Free PMC article. - Regulation of gene expression through a transcriptional repressor that senses acyl-chain length in membrane phospholipids.
Hofbauer HF, Schopf FH, Schleifer H, Knittelfelder OL, Pieber B, Rechberger GN, Wolinski H, Gaspar ML, Kappe CO, Stadlmann J, Mechtler K, Zenz A, Lohner K, Tehlivets O, Henry SA, Kohlwein SD. Hofbauer HF, et al. Dev Cell. 2014 Jun 23;29(6):729-39. doi: 10.1016/j.devcel.2014.04.025. Dev Cell. 2014. PMID: 24960695 Free PMC article.
References
- Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell. 2000;103:667–678. - PubMed
- Ambroziak J, Henry S A. INO2 and INO4 gene products, positive regulators of phospholipid biosynthesis in Saccharomyces cerevisiae, form a complex that binds to the INO1 promoter. J Biol Chem. 1994;269:15344–15349. - PubMed
- Becker G W, Lester R L. Changes in phospholipids of Saccharomyces cerevisiae associated with inositol-less death. J Biol Chem. 1977;252:8684–8691. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM19629/GM/NIGMS NIH HHS/United States
- AI01816/AI/NIAID NIH HHS/United States
- K02 AI001816/AI/NIAID NIH HHS/United States
- R37 GM019629/GM/NIGMS NIH HHS/United States
- F32 GM019629/GM/NIGMS NIH HHS/United States
- GM52593/GM/NIGMS NIH HHS/United States
- R01 GM052593/GM/NIGMS NIH HHS/United States
- R01 GM019629/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous