Antagonistic controls of autophagy and glycogen accumulation by Snf1p, the yeast homolog of AMP-activated protein kinase, and the cyclin-dependent kinase Pho85p - PubMed (original) (raw)
Antagonistic controls of autophagy and glycogen accumulation by Snf1p, the yeast homolog of AMP-activated protein kinase, and the cyclin-dependent kinase Pho85p
Z Wang et al. Mol Cell Biol. 2001 Sep.
Abstract
In the yeast Saccharomyces cerevisiae, glycogen is accumulated as a carbohydrate reserve when cells are deprived of nutrients. Yeast mutated in SNF1, a gene encoding a protein kinase required for glucose derepression, has diminished glycogen accumulation and concomitant inactivation of glycogen synthase. Restoration of synthesis in an snf1 strain results only in transient glycogen accumulation, implying the existence of other SNF1-dependent controls of glycogen storage. A genetic screen revealed that two genes involved in autophagy, APG1 and APG13, may be regulated by SNF1. Increased autophagic activity was observed in wild-type cells entering the stationary phase, but this induction was impaired in an snf1 strain. Mutants defective for autophagy were able to synthesize glycogen upon approaching the stationary phase, but were unable to maintain their glycogen stores, because subsequent synthesis was impaired and degradation by phosphorylase, Gph1p, was enhanced. Thus, deletion of GPH1 partially reversed the loss of glycogen accumulation in autophagy mutants. Loss of the vacuolar glucosidase, SGA1, also protected glycogen stores, but only very late in the stationary phase. Gph1p and Sga1p may therefore degrade physically distinct pools of glycogen. Pho85p is a cyclin-dependent protein kinase that antagonizes SNF1 control of glycogen synthesis. Induction of autophagy in pho85 mutants entering the stationary phase was exaggerated compared to the level in wild-type cells, but was blocked in apg1 pho85 mutants. We propose that Snf1p and Pho85p are, respectively, positive and negative regulators of autophagy, probably via Apg1 and/or Apg13. Defective glycogen storage in snf1 cells can be attributed to both defective synthesis upon entry into stationary phase and impaired maintenance of glycogen levels caused by the lack of autophagy.
Figures
FIG. 1
Glycogen accumulation in snf1 pcl8 pcl10 cells with multicopy suppressor plasmids. Strains are EG328-1A (wild type [WT]) or ZWS2 (snf1 pcl8 pcl10). The plasmids are pRS425 (vector) or pYEP13 with the indicated gene inserted. In the screen, APG1 was found 17 times, APG13 was found 1 time, PDE2 was found 8 times, RCK2 was found 3 times, GAC1 was found 2 times, and YGR161C was found 2 times. The cells were grown on SD-Leu plates for 2 days at 30°C and then exposed to iodine vapor for 2 min to assess glycogen accumulation, with the darker the staining, the greater the glycogen accumulation.
FIG. 2
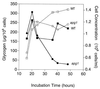
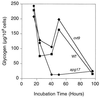
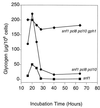
Glycogen accumulation in apg1 cells after prolonged incubation in synthetic complete medium. Wild-type (EG328-1A) and apg1 (ZWP1-5) cells were grown in liquid SD medium. At the indicated times, cell concentrations were measured by counting cell numbers (□, wild-type cells; ○, apg1 cells). Aliquots were taken for determination of glycogen content (■, wild-type cells; ●, apg1 cells), as described as in Materials and Methods. Representative data from one of three independent experiments are shown.
FIG. 3
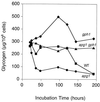
Loss of GPH1 reverses glycogen depletion in apg1 cells. Cells were grown in liquid SD medium, and aliquots were taken at the indicated times for determination of glycogen content as described in Materials and Methods. The following strains were used: EG328-1A (wild type [■]), ZWP1-2 (apg1 [●]) ZW49-161 (apg1 gph1 [▴]), and ZW45-7 (gph1 [⧫]). Representative data from one of three independent experiments are shown.
FIG. 4
Both GPH1 and SGA1 affect glycogen degradation. Cells were grown in liquid SD medium, and aliquots were taken at the indicated times for glycogen measurement. The following strains were used: EG328-1A (wild type [■]), ZW41-6 (sga1 [▾]), ZW48-3 (apg1 sga1 [●]), and ZW48-3 (gph1 sga1 [⧫]). Representative data from one of three independent experiments are shown.
FIG. 5
Glycogen accumulation in cells disrupted for various autophagy genes. Cells were grown in liquid SD medium, and aliquots were taken at the indicated times for determination of glycogen content as described in Materials and Methods. The following strains were used: EG328-1A (wild type [■]), ZWP1-5 (apg1) and ZW74-2 (apg7) (○), ZW75-3 (aut2 [▾]), ZW76-2 (prb1 [▴]), and ZW77-4 (pra1 [⧫]). Glycogen accumulation of other mutant cells has essentially the same kinetics as that in prb1 cells.
FIG. 6
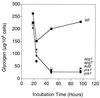
Glycogen accumulation in apg17 and cvt9 cells. The cells were grown in liquid SD medium, and aliquots were taken at the indicated times for determination of glycogen content as described in Materials and Methods. The following strains were used: EG328-1A (wild type [■]), ZW83-1 (apg17 [●]), and ZW84-4 (cvt9 [⧫]). Representative data from one of three independent experiments are shown.
FIG. 7
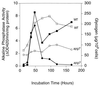
Induction of autophagy upon entry into the stationary phase. Wild-type (TN125) and apg7 (ZW105-3) cells were grown in liquid SD medium. At the indicated times, cells were harvested. Glycogen content (■, wild-type strain TN125 ●, apg7) and alkaline phosphatase activity (□, wild-type strain TN125; ○, apg7) were measured as described in Methods and Materials. Representative data from one of three independent experiments are shown.
FIG. 8
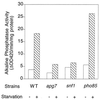
Effect of SNF1 and PHO85 on autophagy. Wild-type (TN125), snf1 (ZW101-3), pho85 (ZW102-1), and apg7 (ZW105-3) cells were grown in YPD to the logarithmic phase before transfer into SD(−N). Incubation was continued for 4 h. Cells were then harvested, and alkaline phosphatase activity was measured as described in Materials and Methods. Representative data from one of three independent experiments are shown.
FIG. 9
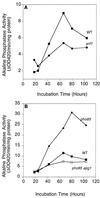
Effects of SNF1 and PHO85 on autophagy upon entry into the stationary phase. (A) Wild-type (TN125) and snf1 (ZW101-3) cells were grown in liquid SD medium. At the indicated times, cells were harvested. Alkaline phosphatase activity (■, wild-type strain TN125 ●, snf1), was measured as described in Materials and Methods. Representative data from one of three independent experiments are shown. (B) Wild-type (TN125), pho85 (ZW102-1), and pho85 apg1 (ZW115-1) cells were grown in liquid SD medium. At the indicated times, cells were harvested. Alkaline phosphatase activity (■, wild-type strain; ⧫, pho85; □, pho85 apg1) was measured as described in Materials and Methods. Representative data from one of three independent experiments are shown.
FIG. 10
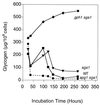
Glycogen accumulation in snf1, snf1 pc18 pcl10, and snf1 pcl8 pcl10 gph1 cells. Cells were grown in YPD overnight and then diluted 1/200 into synthetic complete medium. Samples were collected at the indicated times, and glycogen content was measured as described in Materials and Methods. The following strains were used: EG353-1C (snf1 [■]), ZWS2 (snf1 pcl8 pcl10 [●]), and ZW34-263 (snf1 pcl8 pcl10 gph1 [⧫]). Representative data from one of three independent experiments are shown.
FIG. 11
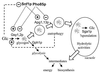
Possible relationships between glycogen metabolism and autophagy. The synthesis and possible fates of glycogen are depicted, with solid arrows representing metabolic interconversion, open arrows indicating physical translocation, and dashed lines indicating regulatory connections. Glycogen is synthesized in the late logarithmic phase through the regulation of glycogen synthase (Gsy1,2p) and other biosynthetic enzymes. At saturation of the culture, there is utilization of glycogen concomitant with activation of phosphorylase (Gph1p) followed by a phase of reaccumulation during which both synthesis and degradation occur simultaneously. Over the same period, starvation signals also lead to increased autophagy, which has at least two effects on glycogen levels. First, it generates, by recycling of cellular components, small molecules such as amino acids that can be returned to the cytosol as a source of energy and/or synthetic intermediates. By an unknown mechanism, autophagy exerts a positive control over glycogen synthesis. A second potential consequence is the sequestration of glycogen in the vacuole, where it is inaccessible to Gph1p, for degradation by the vacuolar glucosidase (Sga1p) very late in the stationary phase. In the absence of autophagy, neither process operates. In the early to mid stationary phase, glycogen is degraded by phosphorylase and is no longer stored in the vacuole, both leading to decreased maintenance of the total glycogen level.
Similar articles
- Pho85p, a cyclin-dependent protein kinase, and the Snf1p protein kinase act antagonistically to control glycogen accumulation in Saccharomyces cerevisiae.
Huang D, Farkas I, Roach PJ. Huang D, et al. Mol Cell Biol. 1996 Aug;16(8):4357-65. doi: 10.1128/MCB.16.8.4357. Mol Cell Biol. 1996. PMID: 8754836 Free PMC article. - Analyses of APG13 gene involved in autophagy in yeast, Saccharomyces cerevisiae.
Funakoshi T, Matsuura A, Noda T, Ohsumi Y. Funakoshi T, et al. Gene. 1997 Jun 19;192(2):207-13. doi: 10.1016/s0378-1119(97)00031-0. Gene. 1997. PMID: 9224892 - SNF1/AMPK pathways in yeast.
Hedbacker K, Carlson M. Hedbacker K, et al. Front Biosci. 2008 Jan 1;13:2408-20. doi: 10.2741/2854. Front Biosci. 2008. PMID: 17981722 Free PMC article. Review. - Reserve carbohydrates metabolism in the yeast Saccharomyces cerevisiae.
François J, Parrou JL. François J, et al. FEMS Microbiol Rev. 2001 Jan;25(1):125-45. doi: 10.1111/j.1574-6976.2001.tb00574.x. FEMS Microbiol Rev. 2001. PMID: 11152943 Review.
Cited by
- The Roles of Ubiquitin in Mediating Autophagy.
Yin Z, Popelka H, Lei Y, Yang Y, Klionsky DJ. Yin Z, et al. Cells. 2020 Sep 2;9(9):2025. doi: 10.3390/cells9092025. Cells. 2020. PMID: 32887506 Free PMC article. Review. - AMPK and Pulmonary Hypertension: Crossroads Between Vasoconstriction and Vascular Remodeling.
Zhao Q, Song P, Zou MH. Zhao Q, et al. Front Cell Dev Biol. 2021 Jun 8;9:691585. doi: 10.3389/fcell.2021.691585. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34169079 Free PMC article. Review. - Atg45 is an autophagy receptor for glycogen, a non-preferred cargo of bulk autophagy in yeast.
Isoda T, Takeda E, Hosokawa S, Hotta-Ren S, Ohsumi Y. Isoda T, et al. iScience. 2024 Apr 26;27(6):109810. doi: 10.1016/j.isci.2024.109810. eCollection 2024 Jun 21. iScience. 2024. PMID: 38832010 Free PMC article. - Regulation of the reserve carbohydrate metabolism by alkaline pH and calcium in Neurospora crassa reveals a possible cross-regulation of both signaling pathways.
Virgilio S, Cupertino FB, Ambrosio DL, Bertolini MC. Virgilio S, et al. BMC Genomics. 2017 Jun 9;18(1):457. doi: 10.1186/s12864-017-3832-1. BMC Genomics. 2017. PMID: 28599643 Free PMC article. - Impaired Cellular Energy Metabolism Contributes to Duck-Enteritis-Virus-Induced Autophagy via the AMPK-TSC2-MTOR Signaling Pathway.
Yin H, Zhao L, Li S, Xu L, Wang Y, Chen H. Yin H, et al. Front Cell Infect Microbiol. 2017 Sep 26;7:423. doi: 10.3389/fcimb.2017.00423. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 29018776 Free PMC article.
References
- Andrews B, Measday V. The cyclin family of budding yeast: abundant use of a good idea. Trends Genet. 1998;14:66–72. - PubMed
- Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. - PubMed
- Carlson M. Glucose repression in yeast. Curr Opin Microbiol. 1999;2:202–207. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases