Junctional adhesion molecule (JAM) binds to PAR-3: a possible mechanism for the recruitment of PAR-3 to tight junctions - PubMed (original) (raw)
Junctional adhesion molecule (JAM) binds to PAR-3: a possible mechanism for the recruitment of PAR-3 to tight junctions
M Itoh et al. J Cell Biol. 2001.
Abstract
At tight junctions (TJs), claudins with four transmembrane domains are incorporated into TJ strands. Junctional adhesion molecule (JAM), which belongs to the immunoglobulin superfamily, is also localized at TJs, but it remains unclear how JAM is integrated into TJs. Immunoreplica electron microscopy revealed that JAM showed an intimate spatial relationship with TJ strands in epithelial cells. In L fibroblasts expressing exogenous JAM, JAM was concentrated at cell-cell adhesion sites, where there were no strand-like structures, but rather characteristic membrane domains free of intramembranous particles were detected. These domains were specifically labeled with anti-JAM polyclonal antibody, suggesting that JAM forms planar aggregates through their lateral self-association. Immunofluorescence microscopy and in vitro binding assays revealed that ZO-1 directly binds to the COOH termini of claudins and JAM at its PDZ1 and PDZ3 domains, respectively. Furthermore, another PDZ-containing polarity-related protein, PAR-3, was directly bound to the COOH terminus of JAM, but not to that of claudins. These findings led to a molecular architectural model for TJs: small aggregates of JAM are tethered to claudin-based strands through ZO-1, and these JAM aggregates recruit PAR-3 to TJs. We also discuss the importance of this model from the perspective of the general molecular mechanisms behind the recruitment of PAR proteins to plasma membranes.
Figures
Figure 1.
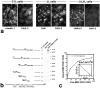
JAM in TJs of epithelial cells. (a and b) Double immunofluorescence staining of cultured MDCK cells with anti-JAM pAb (C4) (a) and anti–ZO-1 mAb (b). JAM was precisely colocalized with ZO-1 at cell–cell adhesion sites. (Bottom) Show vertical-sectional views and their merged view generated from confocal images. Arrowhead, apical level; arrow, basal level. (c and d) Freeze fracture replicas obtained from cultured MDCK cells were immunolabeled with anti-JAM pAb (C-tail). The TJ region was specifically labeled with immunogold particles (c), and most immunogold particles showed an intimate spatial relationship with TJ strands (d). (c and d) The TJ region on the E-face and P-face, respectively, was labeled. The E-face–associated labeling is not easily explained according to Fujimoto's original paper (1995), but similar labeling was observed with anticlaudin and antioccludin pAbs, suggesting that this type of labeling was characteristic to TJ constituents, i.e., that JAM molecules are tightly associated with TJ strands. Mv, microvilli. Bars, 100 nm.
Figure 2.
Lateral aggregation of JAM in L transfectants. (a) Immunofluorescence microscopy of JL cells with anti-JAM pAb (C4). JAM was concentrated at cell–cell adhesion sites as planes. (b) Freeze fracture replica image of cell–cell adhesion sites of JL cells. Characteristic intramembranous particle-free domains (*) were frequently observed. (c) The particle-free domains of JL cells were specifically labeled with pAb-specific for the cytoplasmic domain of JAM (anti-JAM pAb, C-tail). Bar, 200 nm.
Figure 3.
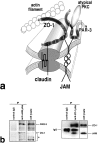
Interaction between JAM and ZO-1. (a) Recruitment of endogenous ZO-1 to cell–cell adhesion sites in L transfectants. C1L cells, JL cells, or JAM lacking its COOH-terminal –LV (JΔLVL cells) were double stained. Claudin-1, JAM, and JAMΔLV were all concentrated at cell–cell adhesion sites. Claudin-1 and JAM, but not JAMΔLV, recruited ZO-1 to cell–cell contact sites (arrowheads). (b) Eight distinct portions of ZO-1 were produced as recombinant fusion proteins with maltose-binding protein (MBP) in E. coli. Their crude lysates containing recombinant proteins (E. coli lysate) were mixed with beads conjugated with GST or GST fusion protein with the cytoplasmic domain of JAM (GST-JAMcyt). Bound proteins were then eluted from GST-conjugated beads (GST eluate) or GST-JAMcyt–conjugated beads (GST-JAMcyt eluate), and each eluate was subjected to SDS-PAGE followed by Coomassie brilliant blue staining. Among eight types of MBP fusion proteins, only MBP–NZO-1, MBP-PDZ2-GUK, and MBP-PDZ3-GUK were bound to GST-JAMcyt. (c) Quantitative analysis of the binding between MBP-NZO-1 and GST-JAMcyt. Glutathione–Sepharose bead slurry containing GST-JAMcyt was incubated with E. coli lysate containing various amounts of MBP–NZO-1. The amounts of MBP–NZO-1 in the E. coli lysate and each eluate (inset) were estimated by comparing the Coomassie Brilliant blue staining intensity of bands. The binding was saturable, and Scatchard analysis (inset) indicated that the K d value was 1.1 × 10−7 M.
Figure 4.
Interaction between JAM and PAR-3. (a) Recruitment of endogenous PAR-3 to cell–cell adhesion sites in L transfectants. C1L cells, JL cells, or JΔLVL cells were double stained. Claudin-1, JAM, and JAMΔLV were all concentrated at cell–cell adhesion sites. JAM, but not claudin-1–JAMΔLV, recruited PAR-3 to cell–cell contact sites (arrowheads). (b) Six distinct portions of PAR-3 were produced as recombinant fusion proteins with MBP in E. coli, and then the same in vitro binding analysis as described in the legend to Fig. 3 b was performed. Among six types of MBP fusion proteins, only MBP–PDZ1-PDZ3 was bound to GST-JAMcyt. (c) Quantitative analysis of the binding between MBP–PDZ1-PDZ3 of PAR-3 and GST-JAMcyt. K d value was determined to be 7.5 × 10−8 M.
Figure 5.
Molecular interactions within TJs in epithelial cells. (a) A model for the molecular architecture of TJs. See details in the text. COOH-terminal domain of ZO-1 is associated with actin filaments. ZO-2 and ZO-3 may also function as cross-linkers like ZO-1 (Itoh et al., 1999), and occludin in TJ strands may also be cross-linked to JAM by ZO-1, although these were not depicted in this model. (b) Immunoprecipitation. As mAb specific for the extracellular domain of JAM recognizes human JAM, but not mouse JAM, human T84 cells were used. (Left) JAM was immunoprecipitated from T84 cell lysate with anti-JAM mAb. Immunoblotting with anti–PAR-3/ASIP pAb or anti–ZO-1 mAb detected PAR-3 or ZO-1, respectively, in the JAM immunoprecipitate. *Degradation products of ZO-1. (Right) ZO-1 was immunoprecipitated with anti–ZO-1 pAb. Immunoblotting with anti-JAM pAb detected JAM in the ZO-1 immunoprecipitate. PAR-3 was undetectable under the condition used in this experiment, but this would be reasonable since PAR-3 is not directly associated with ZO-1.
Similar articles
- Involvement of nectin in the localization of junctional adhesion molecule at tight junctions.
Fukuhara A, Irie K, Nakanishi H, Takekuni K, Kawakatsu T, Ikeda W, Yamada A, Katata T, Honda T, Sato T, Shimizu K, Ozaki H, Horiuchi H, Kita T, Takai Y. Fukuhara A, et al. Oncogene. 2002 Oct 31;21(50):7642-55. doi: 10.1038/sj.onc.1205875. Oncogene. 2002. PMID: 12400007 - The junctional adhesion molecule (JAM) family members JAM-2 and JAM-3 associate with the cell polarity protein PAR-3: a possible role for JAMs in endothelial cell polarity.
Ebnet K, Aurrand-Lions M, Kuhn A, Kiefer F, Butz S, Zander K, Meyer zu Brickwedde MK, Suzuki A, Imhof BA, Vestweber D. Ebnet K, et al. J Cell Sci. 2003 Oct 1;116(Pt 19):3879-91. doi: 10.1242/jcs.00704. J Cell Sci. 2003. PMID: 12953056 - ZO-1 and ZO-2 independently determine where claudins are polymerized in tight-junction strand formation.
Umeda K, Ikenouchi J, Katahira-Tayama S, Furuse K, Sasaki H, Nakayama M, Matsui T, Tsukita S, Furuse M, Tsukita S. Umeda K, et al. Cell. 2006 Aug 25;126(4):741-54. doi: 10.1016/j.cell.2006.06.043. Cell. 2006. PMID: 16923393 - Tight junction-based epithelial microenvironment and cell proliferation.
Tsukita S, Yamazaki Y, Katsuno T, Tamura A, Tsukita S. Tsukita S, et al. Oncogene. 2008 Nov 24;27(55):6930-8. doi: 10.1038/onc.2008.344. Oncogene. 2008. PMID: 19029935 Review. - Junctional adhesion molecule 1 (JAM-1).
Naik UP, Eckfeld K. Naik UP, et al. J Biol Regul Homeost Agents. 2003 Oct-Dec;17(4):341-7. J Biol Regul Homeost Agents. 2003. PMID: 15065765 Review.
Cited by
- Relocalization of junctional adhesion molecule A during inflammatory stimulation of brain endothelial cells.
Stamatovic SM, Sladojevic N, Keep RF, Andjelkovic AV. Stamatovic SM, et al. Mol Cell Biol. 2012 Sep;32(17):3414-27. doi: 10.1128/MCB.06678-11. Epub 2012 Jun 25. Mol Cell Biol. 2012. PMID: 22733993 Free PMC article. - Critical role of tight junctions in drug delivery across epithelial and endothelial cell layers.
González-Mariscal L, Nava P, Hernández S. González-Mariscal L, et al. J Membr Biol. 2005 Sep;207(2):55-68. doi: 10.1007/s00232-005-0807-y. J Membr Biol. 2005. PMID: 16477528 Review. - Par3 regulates polarized convergence between APP and BACE1 in hippocampal neurons.
Sun M, Huang C, Wang H, Zhang H. Sun M, et al. Neurobiol Aging. 2019 May;77:87-93. doi: 10.1016/j.neurobiolaging.2019.01.023. Epub 2019 Jan 30. Neurobiol Aging. 2019. PMID: 30784815 Free PMC article. - The protein kinase SIK downregulates the polarity protein Par3.
Vanlandewijck M, Dadras MS, Lomnytska M, Mahzabin T, Lee Miller M, Busch C, Brunak S, Heldin CH, Moustakas A. Vanlandewijck M, et al. Oncotarget. 2017 Dec 31;9(5):5716-5735. doi: 10.18632/oncotarget.23788. eCollection 2018 Jan 19. Oncotarget. 2017. PMID: 29464029 Free PMC article. - Slit diaphragms contain tight junction proteins.
Fukasawa H, Bornheimer S, Kudlicka K, Farquhar MG. Fukasawa H, et al. J Am Soc Nephrol. 2009 Jul;20(7):1491-503. doi: 10.1681/ASN.2008101117. Epub 2009 May 28. J Am Soc Nephrol. 2009. PMID: 19478094 Free PMC article.
References
- Anderson, J.M., and C.M. van Itallie. 1995. Tight junctions and the molecular basis for regulation of paracellular permeability. Am. J. Physiol. 269:G467–G475. - PubMed
- Bazzoni, G., O.M. Martinez-Estrada, F. Mueller, P. Nelboeck, G. Schmid, T. Bartfai, E. Dejana, and M. Brockhaus. 2000. a. Homophilic interaction of junctional adhesion molecule. J. Biol. Chem. 275:30970–30976. - PubMed
- Bazzoni, G., O.M. Martinez-Estrada, F. Orsenigo, M. Cordenonsi, S. Citi, and E. Dejana. 2000. b. Interaction of junctional adhesion molecule with the tight junction components ZO-1, cingulin, and occludin. J. Biol. Chem. 275:20520–20526. - PubMed
- Fujimoto, K. 1995. Freeze-fracture replica electron microscopy combined with SDS digestion for cytochemical labeling of integral membrane proteins. Application to the immunogold labeling of intercellular junctional complexes. J. Cell Sci. 108:3443–3449. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases