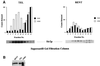Sir2p exists in two nucleosome-binding complexes with distinct deacetylase activities - PubMed (original) (raw)
Sir2p exists in two nucleosome-binding complexes with distinct deacetylase activities
S Ghidelli et al. EMBO J. 2001.
Abstract
The absolute requirement for the histone deacetylase activity of Sir2p in silencing coupled with the conservation of Sir2p-like proteins in larger eukaryotes suggests that this molecule plays an important role in gene regulation in all organisms. Here we report the purification and characterization of two Sir2p-containing protein complexes; one of which contains Sir4p and the other Net1p. The Sir4p-containing complex has an NAD-dependent histone deacetylase activity, while the Net1p-containing complex possesses deacetylase activity but only weak NAD-dependent histone deacetylase activity. Finally, we demonstrate that the Sir2p-containing complexes bind nucleosomes efficiently and partially restrict accessibility of the linker DNA to enzymatic probes.
Figures
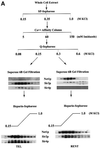
Fig. 1. Sir2p is part of a large stable multi-protein complex in yeast. (A) Elution profile of Sir2p fractionated on a gel-filtration column. A whole cell extract from ROY 1515 was fractionated on a Superose 6B gel-filtration column and 15 µl aliquots of each fraction were analyzed by protein immunoblotting using antisera specific to yeast Sir2p. (B) Partially purified fractions containing Sir2p were treated with either DNase I, RNase A or 500 mM KCl and then fractionated on a Superose 6B gel-filtration column followed by protein immunoblotting using antisera specific to yeast Sir2p.
Fig. 2. Purification of Sir protein complexes from yeast whole cell extracts. (A) The fractionation scheme of Sir2p containing complexes from yeast whole cell extracts is summarized in the flow chart. The immunoblots depict selected fractions from the columns probed with antisera against Sir2p, Sir4p and Net1p. (B) Polypeptide composition of the TEL and RENT complexes. Aliquots of the peak fractions from the heparin-Sepharose column were resolved on a 3–18% SDS–polyacrylamide gel and stained with silver. The asterisks denote bands that co-fractionate with Sir2p. Identities of Sir2p, Sir4p and Net1p were ascertained by protein immunoblotting with antibodies specific to these proteins. (C) Co-immunoprecipitation analysis of Sir4p and Net1p. Whole cell extracts (2 mg) generated from ROY 1914 were subjected to immunoprecipitation with commercial antibodies against the Myc and HA epitopes. The input extract as well as aliquots of the immunoprecipitated proteins were resolved on an SDS–polyacrylamide gel and analyzed by protein immunoblotting with antibodies that recognize Sir2p (lanes 1–4), Sir3p (lanes 5–8), Sir4p (lanes 9–12) and Net1p (lanes 13–16). (D) The fractionation scheme of Sir3p containing complexes from yeast whole cell extracts is summarized in the flow chart. The immunoblots depict selected fractions from the SP-Sepharose column probed with antisera against Sir1p, Sir2p Sir3p and Sir4p.
Fig. 2. Purification of Sir protein complexes from yeast whole cell extracts. (A) The fractionation scheme of Sir2p containing complexes from yeast whole cell extracts is summarized in the flow chart. The immunoblots depict selected fractions from the columns probed with antisera against Sir2p, Sir4p and Net1p. (B) Polypeptide composition of the TEL and RENT complexes. Aliquots of the peak fractions from the heparin-Sepharose column were resolved on a 3–18% SDS–polyacrylamide gel and stained with silver. The asterisks denote bands that co-fractionate with Sir2p. Identities of Sir2p, Sir4p and Net1p were ascertained by protein immunoblotting with antibodies specific to these proteins. (C) Co-immunoprecipitation analysis of Sir4p and Net1p. Whole cell extracts (2 mg) generated from ROY 1914 were subjected to immunoprecipitation with commercial antibodies against the Myc and HA epitopes. The input extract as well as aliquots of the immunoprecipitated proteins were resolved on an SDS–polyacrylamide gel and analyzed by protein immunoblotting with antibodies that recognize Sir2p (lanes 1–4), Sir3p (lanes 5–8), Sir4p (lanes 9–12) and Net1p (lanes 13–16). (D) The fractionation scheme of Sir3p containing complexes from yeast whole cell extracts is summarized in the flow chart. The immunoblots depict selected fractions from the SP-Sepharose column probed with antisera against Sir1p, Sir2p Sir3p and Sir4p.
Fig. 2. Purification of Sir protein complexes from yeast whole cell extracts. (A) The fractionation scheme of Sir2p containing complexes from yeast whole cell extracts is summarized in the flow chart. The immunoblots depict selected fractions from the columns probed with antisera against Sir2p, Sir4p and Net1p. (B) Polypeptide composition of the TEL and RENT complexes. Aliquots of the peak fractions from the heparin-Sepharose column were resolved on a 3–18% SDS–polyacrylamide gel and stained with silver. The asterisks denote bands that co-fractionate with Sir2p. Identities of Sir2p, Sir4p and Net1p were ascertained by protein immunoblotting with antibodies specific to these proteins. (C) Co-immunoprecipitation analysis of Sir4p and Net1p. Whole cell extracts (2 mg) generated from ROY 1914 were subjected to immunoprecipitation with commercial antibodies against the Myc and HA epitopes. The input extract as well as aliquots of the immunoprecipitated proteins were resolved on an SDS–polyacrylamide gel and analyzed by protein immunoblotting with antibodies that recognize Sir2p (lanes 1–4), Sir3p (lanes 5–8), Sir4p (lanes 9–12) and Net1p (lanes 13–16). (D) The fractionation scheme of Sir3p containing complexes from yeast whole cell extracts is summarized in the flow chart. The immunoblots depict selected fractions from the SP-Sepharose column probed with antisera against Sir1p, Sir2p Sir3p and Sir4p.
Fig. 2. Purification of Sir protein complexes from yeast whole cell extracts. (A) The fractionation scheme of Sir2p containing complexes from yeast whole cell extracts is summarized in the flow chart. The immunoblots depict selected fractions from the columns probed with antisera against Sir2p, Sir4p and Net1p. (B) Polypeptide composition of the TEL and RENT complexes. Aliquots of the peak fractions from the heparin-Sepharose column were resolved on a 3–18% SDS–polyacrylamide gel and stained with silver. The asterisks denote bands that co-fractionate with Sir2p. Identities of Sir2p, Sir4p and Net1p were ascertained by protein immunoblotting with antibodies specific to these proteins. (C) Co-immunoprecipitation analysis of Sir4p and Net1p. Whole cell extracts (2 mg) generated from ROY 1914 were subjected to immunoprecipitation with commercial antibodies against the Myc and HA epitopes. The input extract as well as aliquots of the immunoprecipitated proteins were resolved on an SDS–polyacrylamide gel and analyzed by protein immunoblotting with antibodies that recognize Sir2p (lanes 1–4), Sir3p (lanes 5–8), Sir4p (lanes 9–12) and Net1p (lanes 13–16). (D) The fractionation scheme of Sir3p containing complexes from yeast whole cell extracts is summarized in the flow chart. The immunoblots depict selected fractions from the SP-Sepharose column probed with antisera against Sir1p, Sir2p Sir3p and Sir4p.
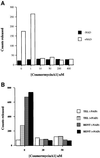
Fig. 3. Analysis of the NAD-dependent histone deacetylase activity. (A) Coumermycin A1 sensitivity of recombinant Sir2p. Deacetylase assays were performed as described using 1 µg of acetylated histones and 0.5 µg of recombinant Sir2p for 30 min at 30°C with varying concentrations of coumermycin A1. The amount of acetate released is depicted in the graph in the presence or absence of added NAD. (B) Coumermycin A1 sensitivity of the TEL and RENT complexes. Deacetylase assays were performed as described using 1 µg of acetylated histones and approximately equal amounts of Sir2p present in the TEL and RENT complexes for 30 min at 30°C with varying concentrations of coumermycin A1. The amount of acetate released is depicted in the graph in the presence or absence of added NAD.
Fig. 4. Analysis of the deacetylase activity of purified Sir2p containing complexes. (A) Co-purification of Sir2p and NAD-dependent deacetylase activity from yeast extracts. Aliquots of the indicated fractions across the Superose 6B gel-filtration column were assayed for NAD-dependent deacetylase activity. A fixed volume of the same fractions was also assayed by protein immunoblotting with antisera against Sir2p. The immunoblot panels are the same as in Figure 2A. (B) Protein immunoblot analysis of Sir2p present in the TEL and RENT complexes. Protein immunoblot analysis with anti-Sir2p antibodies was used to determine approximately equal amounts of Sir2p in the TEL and RENT complexes. Ten microliters (10 µg) of the TEL complex and 40 µl (40 µg) of the RENT complex gave approximately equivalent amounts of signal (equal to ∼0.1 µg of rSir2p). (C) Time course of deacetylation with purified TEL and RENT complexes. Approximately equal amounts of Sir2p present in the TEL and RENT complexes were assayed for deacetylase activity with 1 µg of acetylated histones for varying lengths of time. The amount of acetate released is indicated in the graph. (D) Effect of NAD concentration on the deacetylase activity of the TEL and RENT complexes. The peak fractions containing the TEL and RENT complexes were assayed for deacetylase activity with approximately equal amounts of Sir2p present in the TEL and RENT complexes and 1 µg of histones for 30 min at 30°C with varying concentrations of NAD in the reaction. The amount of acetate released is shown in the graph. (E) Effect of substrate concentration on the deacetylase activity of the TEL and RENT complexes. The peak fractions containing the TEL and RENT complexes were assayed for deacetylase activity for 30 min at 30°C and varying amounts of the histones. The amount of acetate released is shown in the graph. (F) Addition of rSir2p to the TEL and RENT complexes. The deacetylase activity of 0.5 µg of recombinant wild-type or the H364Y mutant Sir2p was measured following addition of these proteins to fractions containing the TEL and RENT complexes. The amount of acetate released is shown in the graph in the presence (black bars) or absence (white bars) of NAD.
Fig. 4. Analysis of the deacetylase activity of purified Sir2p containing complexes. (A) Co-purification of Sir2p and NAD-dependent deacetylase activity from yeast extracts. Aliquots of the indicated fractions across the Superose 6B gel-filtration column were assayed for NAD-dependent deacetylase activity. A fixed volume of the same fractions was also assayed by protein immunoblotting with antisera against Sir2p. The immunoblot panels are the same as in Figure 2A. (B) Protein immunoblot analysis of Sir2p present in the TEL and RENT complexes. Protein immunoblot analysis with anti-Sir2p antibodies was used to determine approximately equal amounts of Sir2p in the TEL and RENT complexes. Ten microliters (10 µg) of the TEL complex and 40 µl (40 µg) of the RENT complex gave approximately equivalent amounts of signal (equal to ∼0.1 µg of rSir2p). (C) Time course of deacetylation with purified TEL and RENT complexes. Approximately equal amounts of Sir2p present in the TEL and RENT complexes were assayed for deacetylase activity with 1 µg of acetylated histones for varying lengths of time. The amount of acetate released is indicated in the graph. (D) Effect of NAD concentration on the deacetylase activity of the TEL and RENT complexes. The peak fractions containing the TEL and RENT complexes were assayed for deacetylase activity with approximately equal amounts of Sir2p present in the TEL and RENT complexes and 1 µg of histones for 30 min at 30°C with varying concentrations of NAD in the reaction. The amount of acetate released is shown in the graph. (E) Effect of substrate concentration on the deacetylase activity of the TEL and RENT complexes. The peak fractions containing the TEL and RENT complexes were assayed for deacetylase activity for 30 min at 30°C and varying amounts of the histones. The amount of acetate released is shown in the graph. (F) Addition of rSir2p to the TEL and RENT complexes. The deacetylase activity of 0.5 µg of recombinant wild-type or the H364Y mutant Sir2p was measured following addition of these proteins to fractions containing the TEL and RENT complexes. The amount of acetate released is shown in the graph in the presence (black bars) or absence (white bars) of NAD.
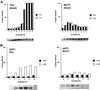
Fig. 5. Purification and analysis of the deacetylase activity of Sir2p-containing complexes purified from mutant cells. Aliquots of the indicated fractions across the gel-filtration column were assayed for NAD-dependent deacetylase activity. (A) The amount of deacetylation mediated by Sir2p containing complexes purified from cells with a mutant Sir2p (H364Y) are shown in graphical form. The immunoblot profile under the graphs is for Sir2p. (B) The amount of deacetylation mediated by Sir2p-containing complexes purified from cells lacking Rpd3p is shown. The immunoblot profile under the graphs is for Sir2p.
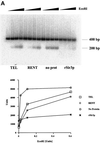
Fig. 6. Sir protein binding to dinucleosomes. (A) Recombinant Sir protein binding to dinucleosomes. Dinucleosomes were reconstituted as described and then bound with varying amounts of recombinant Sir3p or Sir2p. Following binding, the complexes were resolved on a 0.7% agarose gel, the gels were dried and the bands visualized by autoradiography. (B) Reconstitution of mononucleosomes. Mononucleosomes were reconstituted onto a 200 bp fragment of DNA containing a 5S rRNA gene using the octamer exchange method. Increasing amounts of unlabeled mononucleosomes were added to a fixed amount of end-labeled DNA at high salt and the salt concentration was gradually reduced by dilution resulting in the transfer of octamers from unlabeled to labeled DNA fragments. The reconstituted nucleosomes were analyzed by gel mobility shift analysis on a 1% agarose gel in 0.5× TBE. The reaction marked by the asterisk was used for the gel mobility shift data presented in (C). (C) Binding of purified TEL and RENT complexes to mononucleosomes. Reconstituted mononucleosomes were bound with increasing amounts of the purified TEL and RENT complexes and the tertiary complexes were resolved on a 1% agarose gel in 0.4× TBE. (D) Reconstitution of dinucleosomes. Dinucleosomes were reconstituted onto a 400 bp fragment of DNA containing two tandemly repeated 5S rRNA genes using the octamer exchange method as described in (B). The reaction marked by the asterisk was used for the gel mobility shift data presented in (E). (E) Binding of purified TEL and RENT complexes to dinucleosomes. Reconstituted dinucleosomes were bound with increasing amounts of the purified TEL and RENT complexes and the tertiary complexes were resolved on a 0.7% agarose gel in 0.4× TBE.
Fig. 7. Nuclease accessibility analysis of the reconstituted dinucleosomes. (A) Restriction enzyme accessibility analysis of the reconstituted dinucleosomes. Dinucleosomes bound with various Sir protein complexes (lanes 1–4 contained the TEL complex; lanes 5–8 contained the RENT complex; lanes 9–12 had no protein complex added; and lanes 13–16 contained recombinant Sir3p) were digested with increasing concentrations of _Eco_RI (0–0.6 U) for 15 min. The digestion was stopped, DNA was isolated and analyzed on a 1.5% agarose gel followed by autoradiography. The amount of radioactivity in the 200 bp DNA fragment was quantitated and is presented in graphical format. (B) Micrococcal nuclease accessibility analysis of the reconstituted dinucleosomes. Dinucleosomes bound with various Sir protein complexes (lanes 1–4 contained the TEL complex; lanes 5–8 contained the RENT complex; lanes 9–12 had no protein complex added; and lanes 13–16 contained recombinant Sir3p) were digested with increasing concentrations of micrococcal nuclease (0–0.25 U) for 15 min. The samples were then analyzed as described in (A).
Fig. 7. Nuclease accessibility analysis of the reconstituted dinucleosomes. (A) Restriction enzyme accessibility analysis of the reconstituted dinucleosomes. Dinucleosomes bound with various Sir protein complexes (lanes 1–4 contained the TEL complex; lanes 5–8 contained the RENT complex; lanes 9–12 had no protein complex added; and lanes 13–16 contained recombinant Sir3p) were digested with increasing concentrations of _Eco_RI (0–0.6 U) for 15 min. The digestion was stopped, DNA was isolated and analyzed on a 1.5% agarose gel followed by autoradiography. The amount of radioactivity in the 200 bp DNA fragment was quantitated and is presented in graphical format. (B) Micrococcal nuclease accessibility analysis of the reconstituted dinucleosomes. Dinucleosomes bound with various Sir protein complexes (lanes 1–4 contained the TEL complex; lanes 5–8 contained the RENT complex; lanes 9–12 had no protein complex added; and lanes 13–16 contained recombinant Sir3p) were digested with increasing concentrations of micrococcal nuclease (0–0.25 U) for 15 min. The samples were then analyzed as described in (A).
Similar articles
- Sound silencing: the Sir2 protein and cellular senescence.
Defossez PA, Lin SJ, McNabb DS. Defossez PA, et al. Bioessays. 2001 Apr;23(4):327-32. doi: 10.1002/bies.1047. Bioessays. 2001. PMID: 11268038 Review. - Locus specificity determinants in the multifunctional yeast silencing protein Sir2.
Cuperus G, Shafaatian R, Shore D. Cuperus G, et al. EMBO J. 2000 Jun 1;19(11):2641-51. doi: 10.1093/emboj/19.11.2641. EMBO J. 2000. PMID: 10835361 Free PMC article. - Structural analyses of Sum1-1p-dependent transcriptionally silent chromatin in Saccharomyces cerevisiae.
Yu Q, Elizondo S, Bi X. Yu Q, et al. J Mol Biol. 2006 Mar 10;356(5):1082-92. doi: 10.1016/j.jmb.2005.11.089. Epub 2005 Dec 20. J Mol Biol. 2006. PMID: 16406069 - Cloning and characterization of two mouse genes with homology to the yeast Sir2 gene.
Yang YH, Chen YH, Zhang CY, Nimmakayalu MA, Ward DC, Weissman S. Yang YH, et al. Genomics. 2000 Nov 1;69(3):355-69. doi: 10.1006/geno.2000.6360. Genomics. 2000. PMID: 11056054 - The Sir2 protein family: A novel deacetylase for gene silencing and more.
Shore D. Shore D. Proc Natl Acad Sci U S A. 2000 Dec 19;97(26):14030-2. doi: 10.1073/pnas.011506198. Proc Natl Acad Sci U S A. 2000. PMID: 11114164 Free PMC article. Review. No abstract available.
Cited by
- Pnc1p-mediated nicotinamide clearance modifies the epigenetic properties of rDNA silencing in Saccharomyces cerevisiae.
McClure JM, Gallo CM, Smith DL Jr, Matecic M, Hontz RD, Buck SW, Racette FG, Smith JS. McClure JM, et al. Genetics. 2008 Oct;180(2):797-810. doi: 10.1534/genetics.108.091090. Epub 2008 Sep 9. Genetics. 2008. PMID: 18780747 Free PMC article. - The molecular topography of silenced chromatin in Saccharomyces cerevisiae.
Thurtle DM, Rine J. Thurtle DM, et al. Genes Dev. 2014 Feb 1;28(3):245-58. doi: 10.1101/gad.230532.113. Genes Dev. 2014. PMID: 24493645 Free PMC article. - Barrier proteins remodel and modify chromatin to restrict silenced domains.
Oki M, Valenzuela L, Chiba T, Ito T, Kamakaka RT. Oki M, et al. Mol Cell Biol. 2004 Mar;24(5):1956-67. doi: 10.1128/MCB.24.5.1956-1967.2004. Mol Cell Biol. 2004. PMID: 14966276 Free PMC article. - A nonhistone protein-protein interaction required for assembly of the SIR complex and silent chromatin.
Rudner AD, Hall BE, Ellenberger T, Moazed D. Rudner AD, et al. Mol Cell Biol. 2005 Jun;25(11):4514-28. doi: 10.1128/MCB.25.11.4514-4528.2005. Mol Cell Biol. 2005. PMID: 15899856 Free PMC article. - Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae.
Rusché LN, Kirchmaier AL, Rine J. Rusché LN, et al. Mol Biol Cell. 2002 Jul;13(7):2207-22. doi: 10.1091/mbc.e02-03-0175. Mol Biol Cell. 2002. PMID: 12134062 Free PMC article.
References
- Aparicio O.M., Billington,B.L. and Gottschling,D.E. (1991) Modifiers of position effect are shared between telomeric and silent mating-type loci in S.cerevisiae. Cell, 66, 1279–1287. - PubMed
- Brachmann C.B., Sherman,J.M., Devine,S.E., Cameron,E.E., Pillus,L. and Boeke,J.D. (1995) The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression and chromosome stability. Genes Dev., 9, 2888–2902. - PubMed
- Braunstein M., Rose,A.B., Holmes,S.G., Allis,C.D. and Broach,J.R. (1993a) Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev., 7, 592–604. - PubMed
- Braunstein M., Rose,A.B., Holmes,S.G., Allis,C.D. and Broach,J.R. (1993b) Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev., 7, 592–604. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases