Discrete protein determinant directs the species-specific adherence of Porphyromonas gingivalis to oral streptococci - PubMed (original) (raw)
Discrete protein determinant directs the species-specific adherence of Porphyromonas gingivalis to oral streptococci
D R Demuth et al. Infect Immun. 2001 Sep.
Abstract
For pathogens to survive in the human oral cavity, they must identify a suitable niche in the complex multispecies biofilm that exists on oral tissues. The periodontal pathogen Porphyromonas gingivalis adheres to Streptococcus gordonii by interacting with a specific region of the streptococcal SspB polypeptide, designated BAR. However, it does not adhere to Streptococcus mutans, which expresses SpaP, a highly conserved homolog of SspB. Comparison of the predicted secondary structure of BAR with the corresponding region of SpaP suggested that the substitution of Asn for Gly1182 and Val for Pro1185 in SspB may confer a unique local structure that is not conserved in SpaP. A synthetic peptide of 26 amino acids that encompassed residues 1167 to 1193 of SspB promoted avid adherence of P. gingivalis, whereas a peptide derived from the region corresponding to BAR in SpaP was inactive. Substitution of Gly1182 and Pro1185 for Asn1182 and Val1185 in SspB by site-specific mutation generated proteins that were predicted to assume an SpaP-like secondary structure, and the purified proteins did not promote P. gingivalis adherence. Furthermore, Enterococcus faecalis strains expressing the site-specific mutants did not support adherence of P. gingivalis cells. In contrast, P. gingivalis adhered efficiently to E. faecalis strains expressing intact SspB or SspB-SpaP chimeric proteins containing BAR. These results suggest that a region of SspB consisting of 26 amino acids is sufficient to mediate the adherence of P. gingivalis to S. gordonii and that the species specificity of adherence arises from its interaction with a discrete structural determinant of SspB that is not conserved in SpaP.
Figures
FIG. 1
Structure of chimeric SspB-SpaP polypeptides Spac4 and Spac5. Spac4 is composed of SspB residues 1 to 1166 fused to residues 1167 to 1500 of SpaP. Spac5 contains residues 1 to 1250 of SspB fused to residues 1251 to 1500 from SpaP. The region of SspB encompassing residues 1167 to 1250 (BAR) mediates the in vitro adherence of P. gingivalis cells to S. gordonii (2). Conserved alanine-rich and proline-rich repetitive domains of SspB and SpaP are labeled HR and PR, respectively.
FIG. 2
(A) Secondary-structure predictions distinguish BAR from the corresponding sequences of SpaP. Regions of the two sequences predicted to assume an α-helical configuration are shown in red; β-sheet is represented by green italics; random coil is indicated in yellow; and β-turns are shown in lowercase black. (B) Sequence similarity of the synthetic BAR and SpaP peptides. Identical residues are indicated with colons, and conservative substitutions are shown with single dots. The predicted secondary structures of the peptides are color coded as described for panel A.
FIG. 3
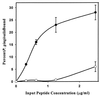
A synthetic peptide comprising residues 1167 to 1193 of BAR promotes adherence of P. gingivalis. Adherence of P. gingivalis to the synthetic BAR (●—●) and SpaP (○—○) peptides was determined by incubating immobilized peptide (0.3 to 2.5 μg/ml) with 3H-labeled P. gingivalis cells (108 total cells) for 2 h with shaking at 25°C. Bound cells were determined by scintillation spectroscopy. Error bars represent standard deviation, n = 3.
FIG. 4
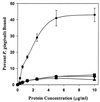
Site-specific mutations in BAR block P. gingivalis adherence to streptococci. Adherence of P. gingivalis to native SspB protein (●—●) and the site-specific mutants N/G1182 (▪—▪), V/P1185 (⧫—⧫), and N/G1182: V/P1185 (▴—▴) was carried out as described previously for the synthetic peptides. Substitution of Pro for Val1185, alone or in combination with a mutation of Gly for Asn1182, resulted in a sequence with a predicted secondary structure identical to that of SpaP. The substitution of Gly for Asn1182 by itself is not predicted to induce a β-turn, but generates a putative α-helix extending to Ser1193. Error bars represent standard deviation, and all experiments were carried out in triplicate.
FIG. 5
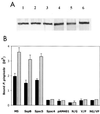
(A) Western blot of SspB polypeptides expressed by E. faecalis SspB, E. faecalis Spac5, E. faecalis Spac4, E. faecalis N/G1182, E. faecalis V/P1185, and E. faecalis NG/VP (lanes 1 to 6, respectively). Protein was obtained by extraction of 109 cells for 5 min in boiling SDS. Samples were electrophoresed on 10% PAGE gels, blotted onto nitrocellulose, and reacted with polyclonal anti-SspB antibodies. (B) Adherence of P. gingivalis to S. gordonii M5 or recombinant E. faecalis strains expressing the SspB, Spac5, or Spac4 protein or the site-specific mutants of SspB. Adherence was determined as described in Materials and Methods using 4 × 107 (black bars) or 8 × 107 (gray bars) input P. gingivalis cells. Error bars represent standard deviation, n = 3.
Similar articles
- Role of the Streptococcus gordonii SspB protein in the development of Porphyromonas gingivalis biofilms on streptococcal substrates.
Lamont RJ, El-Sabaeny A, Park Y, Cook GS, Costerton JW, Demuth DR. Lamont RJ, et al. Microbiology (Reading). 2002 Jun;148(Pt 6):1627-1636. doi: 10.1099/00221287-148-6-1627. Microbiology (Reading). 2002. PMID: 12055284 - Structural characterization of peptide-mediated inhibition of Porphyromonas gingivalis biofilm formation.
Daep CA, James DM, Lamont RJ, Demuth DR. Daep CA, et al. Infect Immun. 2006 Oct;74(10):5756-62. doi: 10.1128/IAI.00813-06. Infect Immun. 2006. PMID: 16988253 Free PMC article. - Identification of a Streptococcus gordonii SspB domain that mediates adhesion to Porphyromonas gingivalis.
Brooks W, Demuth DR, Gil S, Lamont RJ. Brooks W, et al. Infect Immun. 1997 Sep;65(9):3753-8. doi: 10.1128/iai.65.9.3753-3758.1997. Infect Immun. 1997. PMID: 9284148 Free PMC article. - The changing faces of Streptococcus antigen I/II polypeptide family adhesins.
Brady LJ, Maddocks SE, Larson MR, Forsgren N, Persson K, Deivanayagam CC, Jenkinson HF. Brady LJ, et al. Mol Microbiol. 2010 Jul;77(2):276-86. doi: 10.1111/j.1365-2958.2010.07212.x. Epub 2010 May 24. Mol Microbiol. 2010. PMID: 20497507 Free PMC article. Review. - Oral Commensal Streptococci: Gatekeepers of the Oral Cavity.
Baty JJ, Stoner SN, Scoffield JA. Baty JJ, et al. J Bacteriol. 2022 Nov 15;204(11):e0025722. doi: 10.1128/jb.00257-22. Epub 2022 Oct 26. J Bacteriol. 2022. PMID: 36286512 Free PMC article. Review.
Cited by
- Tobacco smoke augments Porphyromonas gingivalis-Streptococcus gordonii biofilm formation.
Bagaitkar J, Daep CA, Patel CK, Renaud DE, Demuth DR, Scott DA. Bagaitkar J, et al. PLoS One. 2011;6(11):e27386. doi: 10.1371/journal.pone.0027386. Epub 2011 Nov 14. PLoS One. 2011. PMID: 22110637 Free PMC article. - A Porphyromonas gingivalis tyrosine phosphatase is a multifunctional regulator of virulence attributes.
Maeda K, Tribble GD, Tucker CM, Anaya C, Shizukuishi S, Lewis JP, Demuth DR, Lamont RJ. Maeda K, et al. Mol Microbiol. 2008 Sep;69(5):1153-64. doi: 10.1111/j.1365-2958.2008.06338.x. Epub 2008 Jun 28. Mol Microbiol. 2008. PMID: 18573179 Free PMC article. - The pathogenic persona of community-associated oral streptococci.
Whitmore SE, Lamont RJ. Whitmore SE, et al. Mol Microbiol. 2011 Jul;81(2):305-14. doi: 10.1111/j.1365-2958.2011.07707.x. Epub 2011 Jun 3. Mol Microbiol. 2011. PMID: 21635580 Free PMC article. Review. - Dendritic cells a critical link to alveolar bone loss and systemic disease risk in periodontitis: Immunotherapeutic implications.
El-Awady AR, Elashiry M, Morandini AC, Meghil MM, Cutler CW. El-Awady AR, et al. Periodontol 2000. 2022 Jun;89(1):41-50. doi: 10.1111/prd.12428. Epub 2022 Mar 4. Periodontol 2000. 2022. PMID: 35244951 Free PMC article. Review. - The Multifaceted Nature of Streptococcal Antigen I/II Proteins in Colonization and Disease Pathogenesis.
Manzer HS, Nobbs AH, Doran KS. Manzer HS, et al. Front Microbiol. 2020 Nov 25;11:602305. doi: 10.3389/fmicb.2020.602305. eCollection 2020. Front Microbiol. 2020. PMID: 33329493 Free PMC article. Review.
References
- Curran T M, Ma Y, Rutherford G C, Marquis R E. Turning on and turning off the arginine deiminase system in oral streptococci. Can J Microbiol Immunol. 1998;44:1078–1085. - PubMed
- Cutler C W, Kalmar J R, Genco C A. Pathogenic strategies of the oral anaerobe Porphyromonas gingivalis. Trends Microbiol. 1995;3:45–51. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- DE12505/DE/NIDCR NIH HHS/United States
- DE12750/DE/NIDCR NIH HHS/United States
- P60 DE013061/DE/NIDCR NIH HHS/United States
- R01 DE012505/DE/NIDCR NIH HHS/United States
- DE13061/DE/NIDCR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources