Candida tropicalis Etr1p and Saccharomyces cerevisiae Ybr026p (Mrf1'p), 2-enoyl thioester reductases essential for mitochondrial respiratory competence - PubMed (original) (raw)
Candida tropicalis Etr1p and Saccharomyces cerevisiae Ybr026p (Mrf1'p), 2-enoyl thioester reductases essential for mitochondrial respiratory competence
J M Torkko et al. Mol Cell Biol. 2001 Sep.
Abstract
We report here on the identification and characterization of novel 2-enoyl thioester reductases of fatty acid metabolism, Etr1p from Candida tropicalis and its homolog Ybr026p (Mrf1'p) from Saccharomyces cerevisiae. Overexpression of these proteins in S. cerevisiae led to the development of significantly enlarged mitochondria, whereas deletion of the S. cerevisiae YBR026c gene resulted in rudimentary mitochondria with decreased contents of cytochromes and a respiration-deficient phenotype. Immunolocalization and in vivo targeting experiments showed these proteins to be predominantly mitochondrial. Mitochondrial targeting was essential for complementation of the mutant phenotype, since targeting of the reductases to other subcellular locations failed to reestablish respiratory growth. The mutant phenotype was also complemented by a mitochondrially targeted FabI protein from Escherichia coli. FabI represents a nonhomologous 2-enoyl-acyl carrier protein reductase that participates in the last step of the type II fatty acid synthesis. This indicated that 2-enoyl thioester reductase activity was critical for the mitochondrial function. We conclude that Etr1p and Ybr026p are novel 2-enoyl thioester reductases required for respiration and the maintenance of the mitochondrial compartment, putatively acting in mitochondrial synthesis of fatty acids.
Figures
FIG. 1
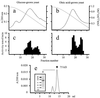
Purification of Etr1p. Soluble extracts (5 mg) from glucose (a)- and oleic acid (b)-grown C. tropicalis were applied to a phenyl-Sepharose column. Bound proteins (solid line) were eluted with a decreasing linear (NH4)2SO4 gradient (dashed line). Respective elution profiles (c and d) of reductase activities determined with _trans_-2,_trans_-4-dienoyl-CoA as substrate are shown. (e) Etr1p (150 μg) from a Mono Q column was run on a Superdex 200 size-exclusion column and eluted at a volume of 13 to 16 ml. A sample from the pooled Etr1p was subjected to SDS-PAGE and Coomassie stained, as shown in the inset.
FIG. 2
Gas chromatographic analysis of the reductase reaction products. Etr1p was incubated with _trans_-2,_trans_-4-hexadienoyl-CoA (a) or _trans_-2-hexenoyl-CoA (b) for 0 (I), 45 (II), 90 (III), and 180 (IV) min in the presence of NADPH, and the hydrolyzed and esterified reaction products were analyzed. After 180 min of incubation with _trans_-2,_trans_-4-hexadienoyl-CoA, _trans_-3-hexenoyl-CoA was added into the reaction mixture, shown in panel a (V). The peaks indicating different ethyl esters derived from CoA esters are _trans-_4-hexenoate (1), _trans_-3-hexenoate (2), _trans_-2,_trans_-4-hexadienoate (3), hexanoate (4), and _trans-_2-hexenoate (5). FID, flame ionization detector.
FIG. 3
Sequence alignment of Etr1p (Etr1_Ct) with mitochondrial respiratory function protein from S. cerevisiae (Mrf1_Sc; accession number D26606), a mitochondrial respiratory function protein homolog from S. pombe (Mrf1_Sp; accession number Q10488), quinone oxidoreductase from E. coli (QOR_Ec; accession number P28304), and ζ-crystallin from Leishmania amazonensis (Zcr_La; accession number L11705). The black and gray boxes indicate identity and similarity, respectively. The amino acid residues are numbered from the initial methionine (+1).
FIG. 4
Comparison of the growth of S. cerevisiae strains on a nonfermentable carbon source. The S. cerevisiae strains BJ1991 wild type, BJ1991 _ybr026c_Δ, or BJ1991 _ybr026c_Δ transformed with plasmid DNA overexpressing either full-length Ybr026p, mature Ybr026p without 5′ leader sequence, Ybr026p extended with a nuclear targeting signal, full-length Etr1p, mature Etr1p without 5′ leader sequence, FabI extended with a mitochondrial targeting signal, or Cta1p were cultured on 3% synthetic complete glycerol medium for 5 days at 30°C.
FIG. 5
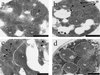
Immuno-EM of Etr1p, Ybr026p, and FabI. C. tropicalis wild type (a) and the S. cerevisiae strains BJ1991 (b), BJ1991 _ybr026c_Δ (c), and BJ1991 _ybr026c_Δ overexpressing either full-length Etr1p (d), full-length Ybr026p (e), mature Etr1p without 5′ leader sequence (f), matureYbr026p without 5′ leader sequence (g), Ybr026p extended with a nuclear targeting signal (h), or FabI extended with a mitochondrial targeting signal (i) were grown on oleic acid. Cells were treated with anti-Etr1p antibodies and gold particles conjugated to protein A. Mitochondria (m) and the nucleus (n) are indicated. The scale bars represent 250 nm.
FIG. 5
Immuno-EM of Etr1p, Ybr026p, and FabI. C. tropicalis wild type (a) and the S. cerevisiae strains BJ1991 (b), BJ1991 _ybr026c_Δ (c), and BJ1991 _ybr026c_Δ overexpressing either full-length Etr1p (d), full-length Ybr026p (e), mature Etr1p without 5′ leader sequence (f), matureYbr026p without 5′ leader sequence (g), Ybr026p extended with a nuclear targeting signal (h), or FabI extended with a mitochondrial targeting signal (i) were grown on oleic acid. Cells were treated with anti-Etr1p antibodies and gold particles conjugated to protein A. Mitochondria (m) and the nucleus (n) are indicated. The scale bars represent 250 nm.
FIG. 5
Immuno-EM of Etr1p, Ybr026p, and FabI. C. tropicalis wild type (a) and the S. cerevisiae strains BJ1991 (b), BJ1991 _ybr026c_Δ (c), and BJ1991 _ybr026c_Δ overexpressing either full-length Etr1p (d), full-length Ybr026p (e), mature Etr1p without 5′ leader sequence (f), matureYbr026p without 5′ leader sequence (g), Ybr026p extended with a nuclear targeting signal (h), or FabI extended with a mitochondrial targeting signal (i) were grown on oleic acid. Cells were treated with anti-Etr1p antibodies and gold particles conjugated to protein A. Mitochondria (m) and the nucleus (n) are indicated. The scale bars represent 250 nm.
FIG. 6
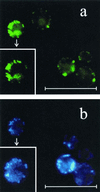
Fluorescent signals from Ybr026p-GFP and DAPI-stained DNA in S. cerevisiae BJ1991. (a) Cells overexpressing Ybr026p-GFP showed punctate green fluorescence. (b) DAPI staining (blue) of the same cells shown in panel a. Punctate staining of mitochondrial DNA was clearly visible and coincided with GFP fluorescence. Additionally, inserts of a single-cell magnification of the corresponding fields of Mrf1p-GFP and DAPI staining are shown. The scale bars represent 16.5 μm.
Similar articles
- Candida tropicalis expresses two mitochondrial 2-enoyl thioester reductases that are able to form both homodimers and heterodimers.
Torkko JM, Koivuranta KT, Kastaniotis AJ, Airenne TT, Glumoff T, Ilves M, Hartig A, Gurvitz A, Hiltunen JK. Torkko JM, et al. J Biol Chem. 2003 Oct 17;278(42):41213-20. doi: 10.1074/jbc.M307664200. Epub 2003 Jul 30. J Biol Chem. 2003. PMID: 12890667 - Structure-function analysis of enoyl thioester reductase involved in mitochondrial maintenance.
Airenne TT, Torkko JM, Van den plas S, Sormunen RT, Kastaniotis AJ, Wierenga RK, Hiltunen JK. Airenne TT, et al. J Mol Biol. 2003 Mar 14;327(1):47-59. doi: 10.1016/s0022-2836(03)00038-x. J Mol Biol. 2003. PMID: 12614607 - Characterization of 2-enoyl thioester reductase from mammals. An ortholog of YBR026p/MRF1'p of the yeast mitochondrial fatty acid synthesis type II.
Miinalainen IJ, Chen ZJ, Torkko JM, Pirilä PL, Sormunen RT, Bergmann U, Qin YM, Hiltunen JK. Miinalainen IJ, et al. J Biol Chem. 2003 May 30;278(22):20154-61. doi: 10.1074/jbc.M302851200. Epub 2003 Mar 24. J Biol Chem. 2003. PMID: 12654921 - Mitochondrial fatty acid synthesis--an adopted set of enzymes making a pathway of major importance for the cellular metabolism.
Hiltunen JK, Chen Z, Haapalainen AM, Wierenga RK, Kastaniotis AJ. Hiltunen JK, et al. Prog Lipid Res. 2010 Jan;49(1):27-45. doi: 10.1016/j.plipres.2009.08.001. Epub 2009 Aug 15. Prog Lipid Res. 2010. PMID: 19686777 Review. - Bacterial Enoyl-Reductases: The Ever-Growing List of Fabs, Their Mechanisms and Inhibition.
Hopf FSM, Roth CD, de Souza EV, Galina L, Czeczot AM, Machado P, Basso LA, Bizarro CV. Hopf FSM, et al. Front Microbiol. 2022 Jun 16;13:891610. doi: 10.3389/fmicb.2022.891610. eCollection 2022. Front Microbiol. 2022. PMID: 35814645 Free PMC article. Review.
Cited by
- Mitochondrial fatty acid synthesis is required for normal mitochondrial morphology and function in Trypanosoma brucei.
Guler JL, Kriegova E, Smith TK, Lukes J, Englund PT. Guler JL, et al. Mol Microbiol. 2008 Mar;67(5):1125-42. doi: 10.1111/j.1365-2958.2008.06112.x. Epub 2008 Jan 23. Mol Microbiol. 2008. PMID: 18221265 Free PMC article. - Myocardial overexpression of Mecr, a gene of mitochondrial FAS II leads to cardiac dysfunction in mouse.
Chen Z, Leskinen H, Liimatta E, Sormunen RT, Miinalainen IJ, Hassinen IE, Hiltunen JK. Chen Z, et al. PLoS One. 2009;4(5):e5589. doi: 10.1371/journal.pone.0005589. Epub 2009 May 18. PLoS One. 2009. PMID: 19440339 Free PMC article. - Phenotypic characterization and comparative transcriptomics of evolved Saccharomyces cerevisiae strains with improved tolerance to lignocellulosic derived inhibitors.
Thompson OA, Hawkins GM, Gorsich SW, Doran-Peterson J. Thompson OA, et al. Biotechnol Biofuels. 2016 Sep 20;9:200. doi: 10.1186/s13068-016-0614-y. eCollection 2016. Biotechnol Biofuels. 2016. PMID: 27679668 Free PMC article. - Dual-Localized Enzymatic Components Constitute the Fatty Acid Synthase Systems in Mitochondria and Plastids.
Guan X, Okazaki Y, Zhang R, Saito K, Nikolau BJ. Guan X, et al. Plant Physiol. 2020 Jun;183(2):517-529. doi: 10.1104/pp.19.01564. Epub 2020 Apr 3. Plant Physiol. 2020. PMID: 32245791 Free PMC article.
References
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1989.
- Baba M, Osumi M. Transmission and scanning electron microscopic examination of intracellular organelles in freeze-substituted Kloeckera and Saccharomyces cerevisiae. J Electron Microsc Tech. 1987;5:249–261.
- Brachmann C B, Davies A, Cost G J, Caputo E, Li J, Hieter P, Boeke J D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases