Cell cycle-dependent recruitment of HDAC-1 correlates with deacetylation of histone H4 on an Rb-E2F target promoter - PubMed (original) (raw)
Cell cycle-dependent recruitment of HDAC-1 correlates with deacetylation of histone H4 on an Rb-E2F target promoter
R Ferreira et al. EMBO Rep. 2001 Sep.
Abstract
The transcription factor E2F, which is a key element in the control of cell proliferation, is repressed by Rb and other pocket proteins in growth-arrested differentiating cells, as well as in proliferating cells when they progress through early G1. It is not known whether similar mechanisms are operative in the two situations. A body of data suggests that E2F repression by pocket proteins involves class I histone deacetylases (HDACs). It has been hypothesized that these enzymes are recruited to E2F target promoters where they deacetylate histones. Here we have tested this hypothesis directly by using formaldehyde cross-linked chromatin immunoprecipitation (XChIP) assays to evaluate HDAC association in living cells. Our data show that a histone deacetylase, HDAC-1, is stably bound to an E2F target promoter during early G1 in proliferating cells and released at the G1-S transition. In addition, our results reveal an inverse correlation between HDAC-1 recruitment and histone H4 acetylation on specific lysines.
Figures
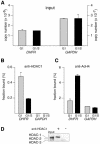
Fig. 1. Cell cycle-dependent recruitment of HDAC-1 and histone H4 deacetylation on DHFR promoter: detection of DHFR promoter in anti-HDAC-1 and anti-acetylated histone H4 immunoprecipitates. Chromatin from synchronized NIH 3T3 cells, either in G1 or at the G1–S transition, as indicated, was analyzed by the XChIP procedure, using anti-HDAC-1 (A and B) or anti-acetylated histone H4 (anti-AcH4) (C and D), followed by PCR analysis of eluted DNA using a LightCycler. The curves show the accumulation of PCR products plotted against the number of cycles (A and C, DHFR; B and D, GAPDH). For the sake of clarity, the results are shown for only one dilution of the immunoprecipitates, but three dilutions were analyzed for each sample. Crosses indicate data points used by the software in calculating copy numbers. (E and F) Curves obtained with reference plasmid DNA for DHFR (E) or GAPDH (F), using 1, 10, 100 and 1000 fg of plasmid.
Fig. 2. Cell cycle-dependent recruitment of HDAC-1 and histone H4 deacetylation on DHFR promoter: compilation of results from Figure 1. Chromatin from synchronized NIH 3T3 cells, either in G1 phase (gray bars) or at the G1–S transition (black bars), was analyzed by the XChIP procedure, followed by quantitative PCR of eluted DNA. Equal amounts of chromatin (A) were subjected to immunoprecipitation using anti-HDAC-1 (B) or anti-AcH4 (C) antibodies. DHFR or GAPDH sequences (as indicated) were detected by quantitative PCR. Copy numbers were estimated by reference to a plasmid containing the promoter sequence and used as a standard; (A) number of copies in the inputs; (B) and (C) fraction of the total number of copies detected as antibody-bound material; a control sample run in parallel in the absence of antibodies revealed little association of DHFR (0.01% of the input) or GAPDH (0.002% of the input). Shown are the results of a typical experiment, with range bars indicating standard deviations of triplicates. This experiment has been reproduced four times with similar results. (D) Chromatin, immunoprecipitated by anti-HDAC-1 (+) or control (–) antibodies or before immunoprecipitation (input), was incubated for 15 min at 95°C and analyzed by western blotting, using an antibody that recognizes HDAC-1, HDAC-2 and HDAC-3 (H80920; Transduction Laboratories).
Fig. 3. Inverse correlation between HDAC-1 recruitment and histone H4 acetylation on DHFR promoter. (A) Chromatin was extracted from cells at different phases of the cell cycle, and immunoprecipitated with anti-HDAC-1 or anti-AcH4 as indicated. The results are expressed as fold variation with reference to the resting cells (mean values from three determinations in three independent experiments, with range bars indicating standard deviations). (B) HDAC-1 protein in immunoprecipitates was monitored in parallel experiments by western blotting with an anti-HDAC antibody. (C) Total RNA was extracted from cells at indicated time points and analyzed by northern blotting using a DHFR or a GAPDH probe, as indicated.
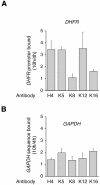
Fig. 4. Lysines 5 and 12 are deacetylated on repressed DHFR promoter. Chromatin was extracted from G1 (4 h) or G1–S (10 h) cells, and immunoprecipitated with anti-AcH4 or anti-acetylated H4 K5, K8, K12 or K16 antibodies, as indicated. The results are expressed as fold variation with reference to the G1 cells (mean values from three determinations in three independent experiments, with range bars indicating standard deviations). (A) DHFR; (B) GAPDH.
Similar articles
- E2F mediates cell cycle-dependent transcriptional repression in vivo by recruitment of an HDAC1/mSin3B corepressor complex.
Rayman JB, Takahashi Y, Indjeian VB, Dannenberg JH, Catchpole S, Watson RJ, te Riele H, Dynlacht BD. Rayman JB, et al. Genes Dev. 2002 Apr 15;16(8):933-47. doi: 10.1101/gad.969202. Genes Dev. 2002. PMID: 11959842 Free PMC article. - Balance between acetylation and methylation of histone H3 lysine 9 on the E2F-responsive dihydrofolate reductase promoter.
Nicolas E, Roumillac C, Trouche D. Nicolas E, et al. Mol Cell Biol. 2003 Mar;23(5):1614-22. doi: 10.1128/MCB.23.5.1614-1622.2003. Mol Cell Biol. 2003. PMID: 12588981 Free PMC article. - The histone deacetylase HDAC3 targets RbAp48 to the retinoblastoma protein.
Nicolas E, Ait-Si-Ali S, Trouche D. Nicolas E, et al. Nucleic Acids Res. 2001 Aug 1;29(15):3131-6. doi: 10.1093/nar/29.15.3131. Nucleic Acids Res. 2001. PMID: 11470869 Free PMC article. - The Rb/chromatin connection and epigenetic control: opinion.
Ferreira R, Naguibneva I, Pritchard LL, Ait-Si-Ali S, Harel-Bellan A. Ferreira R, et al. Oncogene. 2001 May 28;20(24):3128-33. doi: 10.1038/sj.onc.1204337. Oncogene. 2001. PMID: 11420729 Review. - Histone acetylation and the cell-cycle in cancer.
Wang C, Fu M, Mani S, Wadler S, Senderowicz AM, Pestell RG. Wang C, et al. Front Biosci. 2001 Apr 1;6:D610-29. doi: 10.2741/1wang1. Front Biosci. 2001. PMID: 11282573 Review.
Cited by
- G9a promotes proliferation and inhibits cell cycle exit during myogenic differentiation.
Rao VK, Ow JR, Shankar SR, Bharathy N, Manikandan J, Wang Y, Taneja R. Rao VK, et al. Nucleic Acids Res. 2016 Sep 30;44(17):8129-43. doi: 10.1093/nar/gkw483. Epub 2016 May 26. Nucleic Acids Res. 2016. PMID: 27229136 Free PMC article. - Rb/E2F1 regulates the innate immune receptor Toll-like receptor 3 in epithelial cells.
Taura M, Suico MA, Koyama K, Komatsu K, Miyakita R, Matsumoto C, Kudo E, Kariya R, Goto H, Kitajima S, Takahashi C, Shuto T, Nakao M, Okada S, Kai H. Taura M, et al. Mol Cell Biol. 2012 Apr;32(8):1581-90. doi: 10.1128/MCB.06454-11. Epub 2012 Feb 6. Mol Cell Biol. 2012. PMID: 22310660 Free PMC article. - Retinoblastoma family genes.
Du W, Pogoriler J. Du W, et al. Oncogene. 2006 Aug 28;25(38):5190-200. doi: 10.1038/sj.onc.1209651. Oncogene. 2006. PMID: 16936737 Free PMC article. Review. - Rb-E2F-HDAC Repressor Complexes Control Interferon-Induced Repression of Adenovirus To Promote Persistent Infection.
Snaider S, Zheng Y, Hearing P. Snaider S, et al. J Virol. 2022 Jun 8;96(11):e0044222. doi: 10.1128/jvi.00442-22. Epub 2022 May 12. J Virol. 2022. PMID: 35546119 Free PMC article. - TGF-beta induces the expression of SAP30L, a novel nuclear protein.
Lindfors K, Viiri KM, Niittynen M, Heinonen TY, Mäki M, Kainulainen H. Lindfors K, et al. BMC Genomics. 2003 Dec 18;4(1):53. doi: 10.1186/1471-2164-4-53. BMC Genomics. 2003. PMID: 14680513 Free PMC article.
References
- Brehm A., Miska, E.A., McCance, D.J., Reid, J.L., Bannister, A.J. and Kouzarides, T. (1998) Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature, 391, 597–601. - PubMed
- Brehm A., Miska, E., Reid, J., Bannister, A. and Kouzarides, T. (1999) The cell cycle-regulating transcription factors E2F-RB. Br. J. Cancer, 80 Suppl 1, 38–41. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous