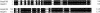The hsp60B gene of Drosophila melanogaster is essential for the spermatid individualization process - PubMed (original) (raw)
The hsp60B gene of Drosophila melanogaster is essential for the spermatid individualization process
B Timakov et al. Cell Stress Chaperones. 2001 Jan.
Abstract
The 60-kDa heat shock protein family (Hsp60) is found in prokaryotes, mitochondria, and chloroplasts. The Hsp60 proteins promote proper protein folding by preventing aggregation. In Drosophila melanogaster, the hsp60 gene is essential for a variety of developmental processes, beginning at early embryogenesis. In this study we show that an additional member of the Drosophila hsp60 gene family, hsp60B, is essential in male fertility. In males homozygous for a mutation of the hsp60B gene, developmental processes appeared normal throughout most of spermatogenesis, including spermatocyte growth, meiosis, and spermatid elongation. At these stages, mitochondria also displayed a differentiation process similar to wild-types. However, we found that the mutation disrupted a late stage of spermatogenesis, the spermatid individualization process. In this process, the individualization complex is assembled at spermatid nuclear heads, traverses along spermatid tails, and generates membranes for each of the spermatids in a cyst. Our analysis further shows that the individualization complex in sterile males displayed abnormal morphology as it was traveling along the spermatid tails. The Drosophila Hsp60 proteins are believed to be exclusively localized in the mitochondria. Our observation that the hsp60B mutation displayed no apparent defect in mitochondrial differentiation during spermatogenesis suggests that the Hsp60B protein may operate in a nonmitochondrial location.
Figures
Fig 1. Alignment of the Hsp60B _N_-terminal sequence with that of Hsp60 (accession number CAA67720). Identical residues are shown as white on black and conserved residues are indicated as black on gray. Dashes show gaps introduced to maximize the alignment. The homology is present along the length of the entire protein sequences
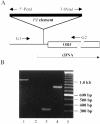
Fig 2. The hsp60B gene is disrupted in the hsp60B1 allele. (A) A schematic drawing of the hsp60B gene structure. The open reading frame and a cDNA sequence of the hsp60B gene are indicated as ORF and cDNA (GenBank accession number AI109018), respectively. _P_-element-specific primers (5′-Pend and 3′-Pend) and _hsp60B_-specific primers (G1 and G2) were employed to show the disruption of the hsp60B gene in the hsp60B1 allele. The arrows represent the primers. (B) Amplification of genomic DNA by PCR. Genomic DNA samples were isolated from wild-type (lane 1) and hsp60B1/hsp60B1 males (lanes 2–4). PCR products were produced from pairs of primers: G1 and G2 (lanes 1 and 2), G1 and 5′-Pend (lane 3), and G2 and 3′-Pend (lane 4). Molecular size standards (100-bp ladder) are shown in lane 5
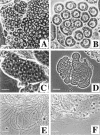
Fig 3. Phase-contrast microscopic analysis of spermatogenesis in hsp60B1/hsp60B1 males. (A) The sizes of primary spermatocytes grew extensively, as in wild-types (from left to right). (B) A cyst of 16 mature primary spermatocytes is shown. (C) A cyst of 64 round spermatids was produced after meiosis. Mitochondria have aggregated to one side of each nucleus (arrow) to form a mitochondrial derivative (arrowhead). (D) A spermatid cyst is shown with elongating tails. (E) At the base of wild-type testes, individual motile sperm with smooth and coiled morphology were produced in large quantity. (F) At the base of hsp60B1/hsp60B1 testes, degrading materials were accumulated. Genotypes: hsp60B1/hsp60B1 (A–D, F) and wild-type (E). Bars, (A) 10 μm; (B–F) 20 μm
Fig 4. The mitochondrial differentiation of spermatogenesis in hsp60B1/hsp60B1 testes stained with MitoTracker Red CMXRos. (A) The mitochondria have aggregated in a cyst of 64 round spermatids after the meiotic divisions. Adjacent to each nucleus (arrow) was a mitochondrial derivative (arrowhead), which was stained selectively by MitoTracker Red CMXRos. (B) The elongating spermatid tails of a 64-cell cyst were stained by MitoTracker Red CMXRos. (C) Fully grown spermatid bundles with 64 tails were stained brightly by the dye. Bars, 10 μm
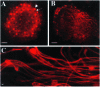
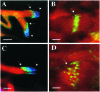
Fig 5. The spermatid individualization process in hsp60B1/hsp60B1 males. In wild-type (A) and hsp60B1/hsp60B1 testes (C), F-actin-rich individualization complexes (green, stained with fluorescein phalloidin, arrow) were assembled onto the aligned nuclear heads (blue, stained with DAPI). (B) In wild-type, an individualization complex with aligned components (stained with fluorescein phalloidin, arrow) traversed along the tails (from right to left). (D) In hsp60B1/hsp60B1 testes, an individualization complex with loosely connected cone- shaped components (stained with fluorescein phalloidin, arrow) traversed along the tails (from right to left). The aligned spermatid tails in bundles were stained with MitoTracker Red CMXRos. Bars, 10 μm
Similar articles
- Testis-Specific Bb8 Is Essential in the Development of Spermatid Mitochondria.
Vedelek V, Laurinyecz B, Kovács AL, Juhász G, Sinka R. Vedelek V, et al. PLoS One. 2016 Aug 16;11(8):e0161289. doi: 10.1371/journal.pone.0161289. eCollection 2016. PLoS One. 2016. PMID: 27529784 Free PMC article. - The Hsp60C gene in the 25F cytogenetic region in Drosophila melanogaster is essential for tracheal development and fertility.
Sarkar S, Lakhotia SC. Sarkar S, et al. J Genet. 2005 Dec;84(3):265-81. doi: 10.1007/BF02715797. J Genet. 2005. PMID: 16385159 - Structure and molecular basis of spermatid elongation in the Drosophila testis.
Huang Q, Chen X, Yu H, Ji L, Shi Y, Cheng X, Chen H, Yu J. Huang Q, et al. Open Biol. 2023 Nov;13(11):230136. doi: 10.1098/rsob.230136. Epub 2023 Nov 8. Open Biol. 2023. PMID: 37935354 Free PMC article. Review. - The tumor suppressor archipelago E3 ligase is required for spermatid differentiation in Drosophila testis.
Vedelek V, Kovács AL, Juhász G, Alzyoud E, Sinka R. Vedelek V, et al. Sci Rep. 2021 Apr 19;11(1):8422. doi: 10.1038/s41598-021-87656-3. Sci Rep. 2021. PMID: 33875704 Free PMC article. - Spermatogenesis in Drosophila. A genetic approach to cellular and subcellular differentiation.
Hackstein JH. Hackstein JH. Eur J Cell Biol. 1991 Dec;56(2):151-69. Eur J Cell Biol. 1991. PMID: 1802704 Review.
Cited by
- Molecular Chaperonin HSP60: Current Understanding and Future Prospects.
Singh MK, Shin Y, Han S, Ha J, Tiwari PK, Kim SS, Kang I. Singh MK, et al. Int J Mol Sci. 2024 May 17;25(10):5483. doi: 10.3390/ijms25105483. Int J Mol Sci. 2024. PMID: 38791521 Free PMC article. Review. - COX4-like, a Nuclear-Encoded Mitochondrial Gene Duplicate, Is Essential for Male Fertility in Drosophila melanogaster.
Eslamieh M, Mirsalehi A, Markova DN, Betrán E. Eslamieh M, et al. Genes (Basel). 2022 Feb 25;13(3):424. doi: 10.3390/genes13030424. Genes (Basel). 2022. PMID: 35327978 Free PMC article. - Analysis of Drosophila melanogaster testis transcriptome.
Vedelek V, Bodai L, Grézal G, Kovács B, Boros IM, Laurinyecz B, Sinka R. Vedelek V, et al. BMC Genomics. 2018 Sep 24;19(1):697. doi: 10.1186/s12864-018-5085-z. BMC Genomics. 2018. PMID: 30249207 Free PMC article. - Molecular characteristics of a novel HSP60 gene and its differential expression in Manila clams (Ruditapes philippinarum) under thermal and hypotonic stress.
Ding J, Li J, Yang D, Yang F, Nie H, Huo Z, Yan X. Ding J, et al. Cell Stress Chaperones. 2018 Mar;23(2):179-187. doi: 10.1007/s12192-017-0796-7. Epub 2017 Dec 22. Cell Stress Chaperones. 2018. PMID: 29273967 Free PMC article. - Testis-Specific Bb8 Is Essential in the Development of Spermatid Mitochondria.
Vedelek V, Laurinyecz B, Kovács AL, Juhász G, Sinka R. Vedelek V, et al. PLoS One. 2016 Aug 16;11(8):e0161289. doi: 10.1371/journal.pone.0161289. eCollection 2016. PLoS One. 2016. PMID: 27529784 Free PMC article.
References
- Adams MD, Celniker SE, Holt RA, et al. The Genome Sequence of. Drosophila melanogaster. Science. 2000;287:2185–2179. - PubMed
- Ashburner M 1989 Drosophila: A Laboratory Handbook and Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Castrillon D, Gonczy P, Alexander S, Rawson R, Eberhart C, Viswanathan S, DiNardo S, Wasserman S. Towards a molecular genetic analysis of spermatogenesis in Drosophila melanogaster: characterization of male-sterile mutants generated by single P element mutagenesis. Genetics. 1993;135:489–505. - PMC - PubMed
- Cooley L, Kelley R, Spradling A. Insertional mutagenesis of the Drosophila genome with single P-elements. Science. 1988;239:1121–1128. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous