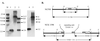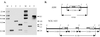Efficient system for directed integration into the Lactobacillus acidophilus and Lactobacillus gasseri chromosomes via homologous recombination - PubMed (original) (raw)
Efficient system for directed integration into the Lactobacillus acidophilus and Lactobacillus gasseri chromosomes via homologous recombination
W M Russell et al. Appl Environ Microbiol. 2001 Sep.
Abstract
An efficient method is described for the generation of site-specific chromosomal integrations in Lactobacillus acidophilus and Lactobacillus gasseri. The strategy is an adaptation of the lactococcal pORI system (K. Leenhouts, G. Venema, and J. Kok, Methods Cell Sci. 20:35-50, 1998) and relies on the simultaneous use of two plasmids. The functionality of the integration strategy was demonstated by the insertional inactivation of the Lactobacillus acidophilus NCFM lacL gene encoding beta-galactosidase and of the Lactobacillus gasseri ADH gusA gene encoding beta-glucuronidase.
Figures
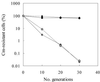
FIG. 1
Stability of pGK12 (♦) and pTRK669 (●) in L. acidophilus NCFM at 37°C (closed symbols) and 43°C (open symbols). The percentage of Cmr cells in each culture was determined by plating on MRS versus MRS plus chloramphenicol.
FIG. 2
(A) Southern hybridization analysis of L. acidophilus NCFM and NCK1398. Chromosomal DNA was digested with _Eco_RI (lanes 1, 2, and 3) or _Hin_dIII (lanes 4 and 5) and hybridized with the 945-bp internal lacL fragment as a probe. Lane 1, pTRK670; lanes 2 and 4, NCFM; lanes 3 and 5, NCK1398; M, digoxigenin-labeled λ-_Hin_dIII marker. DNA sizes are indicated in kilobases. (B) Schematic representation of the relevant regions of the NCFM and NCK1398 chromosomes. Chromosomal DNA is represented by a heavy line, plasmid DNA is represented by a thin line, the lacL gene is represented by an arrow, and the internal lacL fragment is represented by a solid box. The _Eco_RI (E) and _Hin_dIII (H) sites are indicated. The repeating unit represents the plasmid DNA present in various copies (“n”). The diagram is not drawn to scale.
FIG. 3
(A) Southern hybridization analysis of L. gasseri ADH and NCK1423. Chromosomal DNA was digested with _Eco_RI (lanes 1, 2, and 3) or _Eco_RV (lanes 4 and 5) and hybridized with the 777-bp internal gusA fragment as a probe. Lane 1, pTRK685; lanes 2 and 4, ADH; lanes 3 and 5, NCK1423; M, digoxigenin-labeled λ-_Hin_dIII marker. DNA sizes are indicated in kilobases. (B) Schematic representation of the relevant regions of the ADH and NCK1423 chromosomes. Chromosomal DNA is represented by a heavy line, plasmid DNA is represented by a thin line, the gusA gene is represented by an arrow, and the internal gusA fragment is represented by a solid box. The _Eco_RI (E) and _Eco_RV (V) sites are indicated. The diagram is not drawn to scale.
Similar articles
- Construction of an integrative food-grade cloning vector for Lactobacillus acidophilus.
Lin MY, Harlander S, Savaiano D. Lin MY, et al. Appl Microbiol Biotechnol. 1996 May;45(4):484-9. doi: 10.1007/s002530050717. Appl Microbiol Biotechnol. 1996. PMID: 8737572 - Marker-free chromosomal integration of the manganese superoxide dismutase gene (sodA) from Streptococcus thermophilus into Lactobacillus gasseri.
Bruno-Bárcena JM, Azcárate-Peril MA, Klaenhammer TR, Hassan HM. Bruno-Bárcena JM, et al. FEMS Microbiol Lett. 2005 May 1;246(1):91-101. doi: 10.1016/j.femsle.2005.03.044. FEMS Microbiol Lett. 2005. PMID: 15869967 - Directed chromosomal integration and expression of the reporter gene gusA3 in Lactobacillus acidophilus NCFM.
Douglas GL, Klaenhammer TR. Douglas GL, et al. Appl Environ Microbiol. 2011 Oct;77(20):7365-71. doi: 10.1128/AEM.06028-11. Epub 2011 Aug 26. Appl Environ Microbiol. 2011. PMID: 21873486 Free PMC article. - Development and application of a upp-based counterselective gene replacement system for the study of the S-layer protein SlpX of Lactobacillus acidophilus NCFM.
Goh YJ, Azcárate-Peril MA, O'Flaherty S, Durmaz E, Valence F, Jardin J, Lortal S, Klaenhammer TR. Goh YJ, et al. Appl Environ Microbiol. 2009 May;75(10):3093-105. doi: 10.1128/AEM.02502-08. Epub 2009 Mar 20. Appl Environ Microbiol. 2009. PMID: 19304841 Free PMC article. - Invited review: the scientific basis of Lactobacillus acidophilus NCFM functionality as a probiotic.
Sanders ME, Klaenhammer TR. Sanders ME, et al. J Dairy Sci. 2001 Feb;84(2):319-31. doi: 10.3168/jds.S0022-0302(01)74481-5. J Dairy Sci. 2001. PMID: 11233016 Review.
Cited by
- Vaginal Lactobacillus fatty acid response mechanisms reveal a metabolite-targeted strategy for bacterial vaginosis treatment.
Zhu M, Frank MW, Radka CD, Jeanfavre S, Xu J, Tse MW, Pacheco JA, Kim JS, Pierce K, Deik A, Hussain FA, Elsherbini J, Hussain S, Xulu N, Khan N, Pillay V, Mitchell CM, Dong KL, Ndung'u T, Clish CB, Rock CO, Blainey PC, Bloom SM, Kwon DS. Zhu M, et al. Cell. 2024 Sep 19;187(19):5413-5430.e29. doi: 10.1016/j.cell.2024.07.029. Epub 2024 Aug 19. Cell. 2024. PMID: 39163861 Free PMC article. - Assessing immunogenicity of CRISPR-NCas9 engineered strain against porcine epidemic diarrhea virus.
Li F, Zhao H, Sui L, Yin F, Liu X, Guo G, Li J, Jiang Y, Cui W, Shan Z, Zhou H, Wang L, Qiao X, Tang L, Wang X, Li Y. Li F, et al. Appl Microbiol Biotechnol. 2024 Mar 2;108(1):248. doi: 10.1007/s00253-023-12989-0. Appl Microbiol Biotechnol. 2024. PMID: 38430229 Free PMC article. - Vaginal Lactobacillus fatty acid response mechanisms reveal a novel strategy for bacterial vaginosis treatment.
Zhu M, Frank MW, Radka CD, Jeanfavre S, Tse MW, Pacheco JA, Pierce K, Deik A, Xu J, Hussain S, Hussain FA, Xulu N, Khan N, Pillay V, Dong KL, Ndung'u T, Clish CB, Rock CO, Blainey PC, Bloom SM, Kwon DS. Zhu M, et al. bioRxiv [Preprint]. 2023 Dec 30:2023.12.30.573720. doi: 10.1101/2023.12.30.573720. bioRxiv. 2023. PMID: 38234804 Free PMC article. Updated. Preprint. - Recent advances in genetic tools for engineering probiotic lactic acid bacteria.
Mugwanda K, Hamese S, Van Zyl WF, Prinsloo E, Du Plessis M, Dicks LMT, Thimiri Govinda Raj DB. Mugwanda K, et al. Biosci Rep. 2023 Jan 31;43(1):BSR20211299. doi: 10.1042/BSR20211299. Biosci Rep. 2023. PMID: 36597861 Free PMC article. - New Effective Method of Lactococcus Genome Editing Using Guide RNA-Directed Transposition.
Pechenov PY, Garagulya DA, Stanovov DS, Letarov AV. Pechenov PY, et al. Int J Mol Sci. 2022 Nov 12;23(22):13978. doi: 10.3390/ijms232213978. Int J Mol Sci. 2022. PMID: 36430465 Free PMC article.
References
- Kleeman E G, Klaenhammer T R. Adherence of Lactobacillus spp. to human fetal intestinal cells. J Dairy Sci. 1982;65:2063–2069. - PubMed
- Leenhouts K, Bolhuis A, Boot J, Deutz I, Toonen M, Venema G, Kok J, Ledeboer A. Cloning, expression, and chromosomal stabilization of the Propionibacterium shermanii proline iminopeptidase gene (pip) for food-grade application in Lactococcus lactis. Appl Environ Microbiol. 1998;64:4736–4742. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials