Improvement of reading frame maintenance is a common function for several tRNA modifications - PubMed (original) (raw)
Comparative Study
Improvement of reading frame maintenance is a common function for several tRNA modifications
J Urbonavicius et al. EMBO J. 2001.
Abstract
Transfer RNAs from all organisms contain many modified nucleosides. Their vastly different chemical structures, their presence in different tRNAs, their occurrence in different locations in tRNA and their influence on different reactions in which tRNA participates suggest that each modified nucleoside may have its own specific function. However, since the frequency of frameshifting in several different mutants [mnmA, mnmE, tgt, truA (hisT), trmD, miaA, miaB and miaE] defective in tRNA modification was higher compared with the corresponding wild-type controls, these modifications have a common function: they all improve reading frame maintenance. Frameshifting occurs by peptidyl-tRNA slippage, which is influenced by the hypomodified tRNA in two ways: (i) a hypomodified tRNA in the ternary complex may decrease the rate by which the complex is recruited to the A-site and thereby increasing peptidyl-tRNA slippage; or (ii) a hypomodified peptidyl-tRNA may be more prone to slip than its fully modified counterpart. We propose that the improvement of reading frame maintenance has been and is the major selective factor for the emergence of new modified nucleosides.
Figures
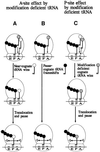
Fig. 1. A dual-error model for frameshifting. (A) Hypomodified cognate tRNA is defective in the aa-tRNA selection step, thereby allowing a wild-type near-cognate tRNA to be accepted instead at the A-site. After a normal three nucleotide translocation, the near-cognate tRNA slips into either –1 or +1 frame, depending on the sequence of the mRNA. (B) The hypomodified cognate tRNA is very slow in entering the A-site, inducing a pause and thereby allowing the wild-type cognate tRNA in the P-site to frameshift. (C) As in (A), but the hypomodified cognate tRNA is accepted in the A-site, and after a normal three nucleotide translocation to the P-site, the hypo modification induces slippage into the –1 or +1 frame. For clarity, only one tRNA is depicted as residing on the ribosome, although two tRNAs are always present in the A- and P-sites or P- and E-sites.
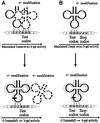
Fig. 2. The assay systems for measuring +1 frameshifting. (A) Measurement of A-site effect by the hypomodified tRNA. The lacZ gene is placed downstream of a short frameshifting window in such a way that β-galactosidase (b-gal) activity is a direct measurement of the frequency with which the ribosome shifts frame within the short frameshifting window. This frequency of frameshifting is dependent on the competition between the peptidyl-tRNA slipping +1 base and aa-tRNA selection at the A-site. The different test codons [NNN in (A)] are placed just downstream of the frameshifting peptidyl-tRNA. (B) Measurement of P-site effect by the hypomodified tRNA. In this case, a stop codon (most often UAG) is placed just downstream of the P-site codon and the lacZ gene is placed in the +1 frame downstream of the A-site stop codon. The P-site frameshifting competes with the release factor activity in the A-site and the β-galactosidase activity is a direct measurement of the efficiency of frameshifting.
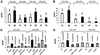
Fig. 3. Influence of the Q34 or mnm5s2U34 on A-site effect by hypomodified tRNA. (A) Influence of Q34 on A-site selection at two His and two Tyr codons. Although the standard error for the His codon CAU suggests a difference between wild type and mutant, the _t_-test analysis did not (Table I). (B) Influence of Q34 on A-site selection at CAU (His), UAU (Tyr) and AAU (Asn) codons. (C) Influence of mnm5s2U34 on A-site selection at two Lys codons. Values obtained using the mnmA mutant were normalized to those using the mnmE mutant. (D) Influence of mnm5s2U34 on two Gln codons. In (A), (C) and (D), the Pro codon CCC is present at the P-site, and in (B) the Phe codon UUU is present at the P-site. Values obtained using the mnmA mutant were normalized to those using the mnmE mutant. The modification status of tRNAs reading A-site codons is indicated above the bars. The frequency of frameshifting is expressed as the β-galactosidase activity normalized to the β-lactamase activity encoded by the bla gene present on the same plasmid as that encoding the lacZ gene.
Fig. 4. Influence of Q34 or mnm5s2U34 tRNA modifications on P-site effect by hypomodified tRNA. (A) Influence of Q34 on slippage at two Tyr codons. (B) Influence of mnm5s2U34 on slippage at two Lys codons. (C) Influence of mnm5s2U34 on slippage at two Gln codons. The stop codon UAG was present at the A-site in all cases. The modification status of tRNA reading P-site codon is indicated above the bars. The frequency of frameshifting is expressed as a percentage of the β-galactosidase activity expressed from the test plasmid compared with that activity of a pseudowild-type β-galactosidase encoded by the control plasmid.
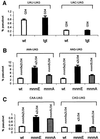
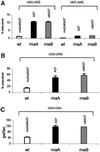
Fig. 5. Influence on P-site effect of ms2io6A37-deficient tRNA. (A) Influence of ms2io6A37 on slippage at two Tyr codons. Stop codon UAG is present at the A-site. (B) As in (A), but the codon UUU is present at the P-site. (C) Influence of ms2io6A37 on slippage at Phe codon UUU. The His codon CAU is present at the A-site. The modification status of tRNA reading P-site codon is indicated above the bars.
Fig. 6. Influence on P-site effect by m1G37-deficient tRNA. (A) Influence of m1G37 on slippage at Leu codons CUU, CUA, CUG and CUC. The value for the CUU codon is 100-fold higher than shown in the figure. (B) Influence of m1G37 on slippage at the Arg codon CGG. (C) Influence of m1G37 on slippage at the Pro codon CCG. In (A) and (B), the stop codon UAG is present in the A-site, and in (C) the stop codon UGA or UAA is present in the A-site. The modification status of tRNA reading P-site codon is indicated above the bars.
Similar articles
- Deficiency of 1-methylguanosine in tRNA from Salmonella typhimurium induces frameshifting by quadruplet translocation.
Hagervall TG, Tuohy TM, Atkins JF, Björk GR. Hagervall TG, et al. J Mol Biol. 1993 Aug 5;232(3):756-65. doi: 10.1006/jmbi.1993.1429. J Mol Biol. 1993. PMID: 7689113 - The ribosomal grip of the peptidyl-tRNA is critical for reading frame maintenance.
Näsvall SJ, Nilsson K, Björk GR. Näsvall SJ, et al. J Mol Biol. 2009 Jan 16;385(2):350-67. doi: 10.1016/j.jmb.2008.10.069. Epub 2008 Nov 3. J Mol Biol. 2009. PMID: 19013179 - Transfer RNA modifications that alter +1 frameshifting in general fail to affect -1 frameshifting.
Urbonavicius J, Stahl G, Durand JM, Ben Salem SN, Qian Q, Farabaugh PJ, Björk GR. Urbonavicius J, et al. RNA. 2003 Jun;9(6):760-8. doi: 10.1261/rna.5210803. RNA. 2003. PMID: 12756333 Free PMC article. - Functions and interplay of the tRNA-binding sites of the ribosome.
Márquez V, Wilson DN, Nierhaus KH. Márquez V, et al. Biochem Soc Trans. 2002 Apr;30(2):133-40. Biochem Soc Trans. 2002. PMID: 12023840 Review. - tRNA's wobble decoding of the genome: 40 years of modification.
Agris PF, Vendeix FA, Graham WD. Agris PF, et al. J Mol Biol. 2007 Feb 9;366(1):1-13. doi: 10.1016/j.jmb.2006.11.046. Epub 2006 Nov 15. J Mol Biol. 2007. PMID: 17187822 Review.
Cited by
- Methylation at position 32 of tRNA catalyzed by TrmJ alters oxidative stress response in Pseudomonas aeruginosa.
Jaroensuk J, Atichartpongkul S, Chionh YH, Wong YH, Liew CW, McBee ME, Thongdee N, Prestwich EG, DeMott MS, Mongkolsuk S, Dedon PC, Lescar J, Fuangthong M. Jaroensuk J, et al. Nucleic Acids Res. 2016 Dec 15;44(22):10834-10848. doi: 10.1093/nar/gkw870. Epub 2016 Sep 28. Nucleic Acids Res. 2016. PMID: 27683218 Free PMC article. - The current riboswitch landscape in Clostridioides difficile.
Badilla Lobo A, Soutourina O, Peltier J. Badilla Lobo A, et al. Microbiology (Reading). 2024 Oct;170(10):001508. doi: 10.1099/mic.0.001508. Microbiology (Reading). 2024. PMID: 39405103 Free PMC article. Review. - Nuclear-encoded factors involved in post-transcriptional processing and modification of mitochondrial tRNAs in human disease.
Powell CA, Nicholls TJ, Minczuk M. Powell CA, et al. Front Genet. 2015 Mar 10;6:79. doi: 10.3389/fgene.2015.00079. eCollection 2015. Front Genet. 2015. PMID: 25806043 Free PMC article. Review. - tRNA properties help shape codon pair preferences in open reading frames.
Buchan JR, Aucott LS, Stansfield I. Buchan JR, et al. Nucleic Acids Res. 2006 Feb 9;34(3):1015-27. doi: 10.1093/nar/gkj488. Print 2006. Nucleic Acids Res. 2006. PMID: 16473853 Free PMC article. - Catalysis by the second class of tRNA(m1G37) methyl transferase requires a conserved proline.
Christian T, Evilia C, Hou YM. Christian T, et al. Biochemistry. 2006 Jun 20;45(24):7463-73. doi: 10.1021/bi0602314. Biochemistry. 2006. PMID: 16768442 Free PMC article.
References
- Barak Z., Lindsley,D. and Gallant,J. (1996) On the mechanism of leftward frameshifting at several hungry codons. J. Mol. Biol., 256, 676–684. - PubMed
- Björk G.R. (1980) A novel link between the biosynthesis of aromatic amino acids and transfer RNA modification in Escherichia coli. J. Mol. Biol., 140, 391–410. - PubMed
- Björk G.R. (1986) Transfer RNA modification in different organisms. Chem. Scripta, 26B, 91–95.
- Björk G.R. (1995) Biosynthesis and function of modified nucleosides in tRNA. In Söll,D. and RajBhandary,U.L. (eds), tRNA: Structure, Biosynthesis and Function. ASM Press, Washington, DC, pp. 165–205.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases