The chromatin remodelling factor Brg-1 interacts with beta-catenin to promote target gene activation - PubMed (original) (raw)
The chromatin remodelling factor Brg-1 interacts with beta-catenin to promote target gene activation
N Barker et al. EMBO J. 2001.
Abstract
Wnt-induced formation of nuclear Tcf-beta-catenin complexes promotes transcriptional activation of target genes involved in cell fate decisions. Inappropriate expression of Tcf target genes resulting from mutational activation of this pathway is also implicated in tumorigenesis. The C-terminus of beta-catenin is indispensable for the transactivation function, which probably reflects the presence of binding sites for essential transcriptional coactivators such as p300/CBP. However, the precise mechanism of transactivation remains unclear. Here we demonstrate an interaction between beta-catenin and Brg-1, a component of mammalian SWI/SNF and Rsc chromatin-remodelling complexes. A functional consequence of reintroduction of Brg-1 into Brg-1-deficient cells is enhanced activity of a Tcf-responsive reporter gene. Consistent with this, stable expression of inactive forms of Brg-1 in colon carcinoma cell lines specifically inhibits expression of endogenous Tcf target genes. In addition, we observe genetic interactions between the Brg-1 and beta-catenin homologues in flies. We conclude that beta-catenin recruits Brg-1 to Tcf target gene promoters, facilitating chromatin remodelling as a prerequisite for transcriptional activation.
Figures
Fig. 1. β-catenin interacts specifically with Brg-1. (A) Schematic representation of the β-catenin domain structure. The N-terminal domain (grey stripes) contains four conserved serine/threonine phosphorylation sites for GSK-3β, which are essential for mediating destruction of free β-catenin. The central domain comprises 12 imperfect repeats of 42 amino acids (denoted Armadillo repeats 1–12; note the presence of an insertion within repeat 10), which are responsible for mediating many of the interactions between β-catenin and its binding partners. The C-terminal domain (shaded grey) contains potent transcriptional activation elements that are essential for the signalling activity of β-catenin. The regions of β-catenin responsible for mediating interaction with other proteins are indicated by curly brackets. (B) Mapping of the Brg-1 domain responsible for mediating interaction with β-catenin. I–IV denote regions of sequence conservation between Brg-1 and Drosophila brm (Khavari et al., 1993). Brg-1 deletion constructs were co-transformed with the β-catenin Arm1–12 bait into the Y190 reporter yeast strain and positive interactions quantified by measuring the activity of a β-galactosidase reporter gene. The asterisk denotes background β-galactosidase activity, as determined by co-transfection of empty prey vector with the β-catenin bait. (C) Mapping of Armadillo repeats mediating interaction with Brg-1. Baits comprising overlapping regions of the β-catenin Armadillo repeats were co-transformed with the Brg-C1 prey plasmid into the Y190 yeast strain and positive interactions quantified by measuring the activity of a β-galactosidase reporter gene.
Fig. 1. β-catenin interacts specifically with Brg-1. (A) Schematic representation of the β-catenin domain structure. The N-terminal domain (grey stripes) contains four conserved serine/threonine phosphorylation sites for GSK-3β, which are essential for mediating destruction of free β-catenin. The central domain comprises 12 imperfect repeats of 42 amino acids (denoted Armadillo repeats 1–12; note the presence of an insertion within repeat 10), which are responsible for mediating many of the interactions between β-catenin and its binding partners. The C-terminal domain (shaded grey) contains potent transcriptional activation elements that are essential for the signalling activity of β-catenin. The regions of β-catenin responsible for mediating interaction with other proteins are indicated by curly brackets. (B) Mapping of the Brg-1 domain responsible for mediating interaction with β-catenin. I–IV denote regions of sequence conservation between Brg-1 and Drosophila brm (Khavari et al., 1993). Brg-1 deletion constructs were co-transformed with the β-catenin Arm1–12 bait into the Y190 reporter yeast strain and positive interactions quantified by measuring the activity of a β-galactosidase reporter gene. The asterisk denotes background β-galactosidase activity, as determined by co-transfection of empty prey vector with the β-catenin bait. (C) Mapping of Armadillo repeats mediating interaction with Brg-1. Baits comprising overlapping regions of the β-catenin Armadillo repeats were co-transformed with the Brg-C1 prey plasmid into the Y190 yeast strain and positive interactions quantified by measuring the activity of a β-galactosidase reporter gene.
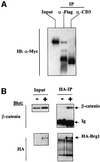
Fig. 2. β-catenin and Brg-1 interact in vivo. (A) 293T cells were transfected with plasmids expressing N-terminal Flag-tagged Brg-C1 clone (amino acids 56–587) and N-terminal Myc-tagged β-catenin. Whole-cell lysates were prepared 24 h later. Extracts were immuno precipitated with anti-Flag antibody or control anti-CD3 antibody as indicated. Precipitated protein was then immunoblotted with anti-Myc antibody to visualize the exogenous tagged β-catenin protein. (B) Expression of full-length K798R Brg-1 protein was induced in DK11 cells by treatment with doxycycline for 24 h. Non-induced (–) and induced (+) cells were then lysed and K798R protein immuno precipitated (IP) with an anti-HA-epitope monoclonal antibody. Precipitated protein was immunoblotted with either anti-HA or anti-β-catenin antibodies. Input lanes show that levels of β-catenin did not differ significantly between non-induced and induced samples, while induction of HA-tagged K798R Brg-1 is clearly visible in lysates before and after immunoprecipitation.
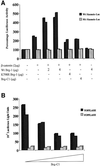
Fig. 3. Brg-1 enhances Tcf–β-catenin transcriptional activity. (A) SW13 cells were transfected with 1 µg of wild-type (black bars) or mutant Siamois reporter plasmid (grey bars) together with the expression constructs indicated. Cells were harvested after 48 h and luciferase activity determined. (B) A partial Brg-1 protein lacking the ATPase domain inhibits constitutive Tcf–β-catenin signalling in DLD1 colon carcinoma cells. A 1 µg aliquot of TOPFLASH (black bars) or FOPFLASH (grey bars) reporter plasmids was transfected into DLD1 cells in the presence or absence of Brg-C1 or full-length Brg-1 expression constructs. Luciferase activities were assayed 48 h later.
Fig. 4. (A) Induction of K798R Brg-1 expression in stable colon carcinoma cell line transfectants. Induction of K798R Brg-1 expression in LS174T (LK1 and LK3) and DLD1 (DK11 and DK19) transfectants by treatment with doxycycline for 24 h was assessed by western blotting using an anti-HA antibody directed against a C-terminal HA tag. (B) Stable expression of dominant-negative Tcf or K798R Brg-1 specifically down-regulates Tcf target genes. Cells with the indicated inducible expression constructs or parental LS174T TR4 and DLD1 TR7 cells were treated with doxycycline (+) or with vehicle alone (–) and RNA isolated 24 h later. RNA, resolved by electrophoresis and transferred to a nylon membrane, was probed with radiolabelled cDNA fragments of the indicated genes.
Fig. 5. Genetic interactions between Brahma complex components and Armadillo in Drosophila. Normal fly eye (A) and wing (F), compared with eyes from GMR.Arm* transformants (B–E) and wings from Engrailed.Gal4 UAS.Cad-I transformants (G–J) in different genetic backgrounds; (B and G) y w; (C and H) _dTC3_2/+; (D and I) _brm_2/+; (E and J) _mor_x/+. Note the strong suppression of the Arm* phenotype in the eye, and the strong enhancement of the Armunder phenotype in the wing due to reduced levels of endogenous Brahma. Similar though less pronounced modifications of the same phenotypes are caused by reducing dTcf and Moira levels.
Similar articles
- The transcriptional coactivator CBP interacts with beta-catenin to activate gene expression.
Takemaru KI, Moon RT. Takemaru KI, et al. J Cell Biol. 2000 Apr 17;149(2):249-54. doi: 10.1083/jcb.149.2.249. J Cell Biol. 2000. PMID: 10769018 Free PMC article. - Interaction and functional cooperation between the LIM protein FHL2, CBP/p300, and beta-catenin.
Labalette C, Renard CA, Neuveut C, Buendia MA, Wei Y. Labalette C, et al. Mol Cell Biol. 2004 Dec;24(24):10689-702. doi: 10.1128/MCB.24.24.10689-10702.2004. Mol Cell Biol. 2004. PMID: 15572674 Free PMC article. - The Yin-Yang of TCF/beta-catenin signaling.
Barker N, Morin PJ, Clevers H. Barker N, et al. Adv Cancer Res. 2000;77:1-24. doi: 10.1016/s0065-230x(08)60783-6. Adv Cancer Res. 2000. PMID: 10549354 Review. - Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma.
Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, Clevers H. Korinek V, et al. Science. 1997 Mar 21;275(5307):1784-7. doi: 10.1126/science.275.5307.1784. Science. 1997. PMID: 9065401 - TCF: Lady Justice casting the final verdict on the outcome of Wnt signalling.
Brantjes H, Barker N, van Es J, Clevers H. Brantjes H, et al. Biol Chem. 2002 Feb;383(2):255-61. doi: 10.1515/BC.2002.027. Biol Chem. 2002. PMID: 11934263 Review.
Cited by
- The WNT/β-catenin dependent transcription: A tissue-specific business.
Söderholm S, Cantù C. Söderholm S, et al. WIREs Mech Dis. 2021 May;13(3):e1511. doi: 10.1002/wsbm.1511. Epub 2020 Oct 21. WIREs Mech Dis. 2021. PMID: 33085215 Free PMC article. Review. - Identification of BCL11B as a regulator of adipogenesis.
Inoue J, Ihara Y, Tsukamoto D, Yasumoto K, Hashidume T, Kamimura K, Nakai Y, Hirano S, Shimizu M, Kominami R, Sato R. Inoue J, et al. Sci Rep. 2016 Sep 2;6:32750. doi: 10.1038/srep32750. Sci Rep. 2016. PMID: 27586877 Free PMC article. - Human SWI/SNF drives sequence-directed repositioning of nucleosomes on C-myc promoter DNA minicircles.
Sims HI, Lane JM, Ulyanova NP, Schnitzler GR. Sims HI, et al. Biochemistry. 2007 Oct 9;46(40):11377-88. doi: 10.1021/bi7008823. Epub 2007 Sep 18. Biochemistry. 2007. PMID: 17877373 Free PMC article. - HMG box transcription factor TCF-4's interaction with CtBP1 controls the expression of the Wnt target Axin2/Conductin in human embryonic kidney cells.
Valenta T, Lukas J, Korinek V. Valenta T, et al. Nucleic Acids Res. 2003 May 1;31(9):2369-80. doi: 10.1093/nar/gkg346. Nucleic Acids Res. 2003. PMID: 12711682 Free PMC article. - Distinct molecular forms of beta-catenin are targeted to adhesive or transcriptional complexes.
Gottardi CJ, Gumbiner BM. Gottardi CJ, et al. J Cell Biol. 2004 Oct 25;167(2):339-49. doi: 10.1083/jcb.200402153. Epub 2004 Oct 18. J Cell Biol. 2004. PMID: 15492040 Free PMC article.
References
- Adams C.C. and Workman,J.L. (1993) Nucleosome displacement in transcription. Cell, 72, 305–308. - PubMed
- Behrens J., von Kries,J.P., Kuhl,M., Bruhn,L., Wedlich,D., Grosschedl,R. and Birchmeier,W. (1996) Functional interaction of β-catenin with the transcription factor LEF-1. Nature, 382, 638–642. - PubMed
- Bienz M. and Clevers,H. (2000) Linking colorectal cancer to Wnt signaling. Cell, 103, 311–320. - PubMed
- Blomquist P., Li,Q. and Wrange,O. (1996) The affinity of nuclear factor 1 for its DNA site is drastically reduced by nucleosome organization irrespective of its rotational or translational position. J. Biol. Chem., 271, 153–159. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous