Hrs recruits clathrin to early endosomes - PubMed (original) (raw)
Hrs recruits clathrin to early endosomes
C Raiborg et al. EMBO J. 2001.
Abstract
The hepatocyte growth factor-regulated tyrosine kinase substrate, Hrs, has been implicated in intracellular trafficking and signal transduction. Hrs contains a phosphatidylinositol 3-phosphate-binding FYVE domain that contributes to its endosomal targeting. Here we show that Hrs and EEA1, a FYVE domain protein involved in endocytic membrane fusion, are localized to different regions of early endosomes. We demonstrate that Hrs co-localizes with clathrin, and that the C-terminus of Hrs contains a functional clathrin box motif that interacts directly with the terminal beta-propeller domain of clathrin heavy chain. A massive recruitment of clathrin to early endosomes was observed in cells transfected with Hrs, but not with Hrs lacking the C-terminus. Furthermore, the phosphatidylinositol 3-kinase inhibitor wortmannin caused the dissociation of both Hrs and clathrin from endosomes. While overexpression of Hrs did not affect endocytosis and recycling of transferrin, endocytosed epidermal growth factor and dextran were retained in early endosomes. These results provide a molecular mechanism for the recruitment of clathrin onto early endosomes and suggest a function for Hrs in trafficking from early to late endosomes.
Figures
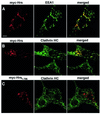
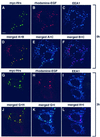
Fig. 1. Localization of Hrs, EEA1 and clathrin in Rab5Q79L-overexpressing BHK cells. BHK cells were transfected with Rab5Q79L and permeabilized with 0.05% saponin prior to fixation. They were then stained with anti-EEA1 (A and C), anti-Hrs (B and E) or anti-clathrin (D and F) and studied by confocal immunofluorescence microscopy. Arrows indicate examples of co-localization. Bar, 5 µm.
Fig. 2. Localization of Hrs, EEA1 and clathrin in melanoma cells. A melanoma cell, permeabilized with 0.05% saponin prior to fixation, was stained with anti-EEA1 (A), anti-Hrs (B) and anti-clathrin (C). Arrows indicate co-localization between EEA1, Hrs and clathrin. Yellow indicates co-localization between Hrs and clathrin (D). The arrowhead points to an endosome that is shown magnified in (G–I). Bar, 5 µm. (E and F) Magnification of the endosomes highlighted by arrows in (A–D). The arrowhead points to an endosome that is shown magnified in (G–I). (G–I) Merged images of a triple-stained magnified endosome. Note the differential localization of EEA1 in blue and Hrs in red, and the co-localization (yellow) between clathrin in green and Hrs.
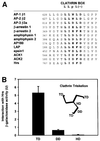
Fig. 3. The C-terminus of Hrs binds clathrin TD. (A) Alignment of sequences found in proteins that bind clathrin (ter Haar et al., 2000; Teo et al., 2001; Yang et al., 2001). This illustrates the existence of a potential clathrin-binding motif within residues 770–775 of Hrs. Hrs is the only protein that has the clathrin box motif at the very C-terminus. (B–E) Interaction of Hrs with clathrin. (B and C) L40 reporter yeast cells were transformed with bait constructs in pLexA and prey constructs in pGAD. Reporter β-galactosidase activities (in arbitrary units) indicate binding and are represented as mean values of two independent experiments performed in duplicate. Error bars denote ± SEM. In (B), a clathrin triskelion (consisting of three heavy chains) is illustrated, with the terminal domain (TD), distal domain (DD) and hub domain (HD) indicated. (D) Recombinant GST (lane 1), GST–Hrs707–775 (lane 2) or GST–Hrs707–770 (lane 3) were immobilized on glutathione–Sepharose beads and incubated with pig brain cytosol. The beads were recovered by centrifugation and washed. Pellet fractions were resolved by SDS–PAGE and transferred to nitrocellulose. The blot was stained with Ponceau S (lower panel) prior to detection of clathrin with anti-clathrin heavy chain antibodies (upper panel). (E) Recombinant GST (lane 1), GST–Hrs707–775 (lane 2) or GST–Hrs707–770 (lane 3), were immobilized on glutathione–Sepharose beads and incubated with purified recombinant clathrin terminal domain (TD1–579). The beads were recovered by centrifugation and washed. Pellet fractions were resolved by SDS–PAGE and transferred to nitrocellulose. The blot was stained with Ponceau S (lower panel) prior to detection of clathrin-TD1–579 with anti-clathrin heavy chain (upper panel). Lane 4 represents the total amount of recombinant clathrin-TD1–579 added to the beads.
Fig. 3. The C-terminus of Hrs binds clathrin TD. (A) Alignment of sequences found in proteins that bind clathrin (ter Haar et al., 2000; Teo et al., 2001; Yang et al., 2001). This illustrates the existence of a potential clathrin-binding motif within residues 770–775 of Hrs. Hrs is the only protein that has the clathrin box motif at the very C-terminus. (B–E) Interaction of Hrs with clathrin. (B and C) L40 reporter yeast cells were transformed with bait constructs in pLexA and prey constructs in pGAD. Reporter β-galactosidase activities (in arbitrary units) indicate binding and are represented as mean values of two independent experiments performed in duplicate. Error bars denote ± SEM. In (B), a clathrin triskelion (consisting of three heavy chains) is illustrated, with the terminal domain (TD), distal domain (DD) and hub domain (HD) indicated. (D) Recombinant GST (lane 1), GST–Hrs707–775 (lane 2) or GST–Hrs707–770 (lane 3) were immobilized on glutathione–Sepharose beads and incubated with pig brain cytosol. The beads were recovered by centrifugation and washed. Pellet fractions were resolved by SDS–PAGE and transferred to nitrocellulose. The blot was stained with Ponceau S (lower panel) prior to detection of clathrin with anti-clathrin heavy chain antibodies (upper panel). (E) Recombinant GST (lane 1), GST–Hrs707–775 (lane 2) or GST–Hrs707–770 (lane 3), were immobilized on glutathione–Sepharose beads and incubated with purified recombinant clathrin terminal domain (TD1–579). The beads were recovered by centrifugation and washed. Pellet fractions were resolved by SDS–PAGE and transferred to nitrocellulose. The blot was stained with Ponceau S (lower panel) prior to detection of clathrin-TD1–579 with anti-clathrin heavy chain (upper panel). Lane 4 represents the total amount of recombinant clathrin-TD1–579 added to the beads.
Fig. 3. The C-terminus of Hrs binds clathrin TD. (A) Alignment of sequences found in proteins that bind clathrin (ter Haar et al., 2000; Teo et al., 2001; Yang et al., 2001). This illustrates the existence of a potential clathrin-binding motif within residues 770–775 of Hrs. Hrs is the only protein that has the clathrin box motif at the very C-terminus. (B–E) Interaction of Hrs with clathrin. (B and C) L40 reporter yeast cells were transformed with bait constructs in pLexA and prey constructs in pGAD. Reporter β-galactosidase activities (in arbitrary units) indicate binding and are represented as mean values of two independent experiments performed in duplicate. Error bars denote ± SEM. In (B), a clathrin triskelion (consisting of three heavy chains) is illustrated, with the terminal domain (TD), distal domain (DD) and hub domain (HD) indicated. (D) Recombinant GST (lane 1), GST–Hrs707–775 (lane 2) or GST–Hrs707–770 (lane 3) were immobilized on glutathione–Sepharose beads and incubated with pig brain cytosol. The beads were recovered by centrifugation and washed. Pellet fractions were resolved by SDS–PAGE and transferred to nitrocellulose. The blot was stained with Ponceau S (lower panel) prior to detection of clathrin with anti-clathrin heavy chain antibodies (upper panel). (E) Recombinant GST (lane 1), GST–Hrs707–775 (lane 2) or GST–Hrs707–770 (lane 3), were immobilized on glutathione–Sepharose beads and incubated with purified recombinant clathrin terminal domain (TD1–579). The beads were recovered by centrifugation and washed. Pellet fractions were resolved by SDS–PAGE and transferred to nitrocellulose. The blot was stained with Ponceau S (lower panel) prior to detection of clathrin-TD1–579 with anti-clathrin heavy chain (upper panel). Lane 4 represents the total amount of recombinant clathrin-TD1–579 added to the beads.
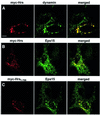
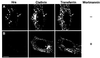
Fig. 4. (A–C) Overexpressed Hrs recruits clathrin to early endosomes. BHK cells were transfected with myc-tagged Hrs (A and B) or Hrs1–706 (C) and permeabilized with 0.05% saponin prior to fixation. They were stained with anti-myc (left panels), anti-EEA1 (A, middle panel) or anti-clathrin (B and C, middle panels) and studied by confocal immunofluorescence microscopy. (D–F) Overexpressed Hrs does not recruit AP1, AP2 or AP3. BHK cells or HEp-2 cells (in the case of AP3) were transfected with myc-tagged Hrs and permeabilized with 0.05% saponin prior to fixation. The cells were stained with anti-myc (left panels), anti-AP1 (D, middle panel), anti-AP2 (E, middle panel) or anti-AP3 (F, middle panel) and studied by confocal immunofluorescence microscopy. The right panels show the merged images, with yellow indicating co-localization. Bar, 5 µm.
Fig. 4. (A–C) Overexpressed Hrs recruits clathrin to early endosomes. BHK cells were transfected with myc-tagged Hrs (A and B) or Hrs1–706 (C) and permeabilized with 0.05% saponin prior to fixation. They were stained with anti-myc (left panels), anti-EEA1 (A, middle panel) or anti-clathrin (B and C, middle panels) and studied by confocal immunofluorescence microscopy. (D–F) Overexpressed Hrs does not recruit AP1, AP2 or AP3. BHK cells or HEp-2 cells (in the case of AP3) were transfected with myc-tagged Hrs and permeabilized with 0.05% saponin prior to fixation. The cells were stained with anti-myc (left panels), anti-AP1 (D, middle panel), anti-AP2 (E, middle panel) or anti-AP3 (F, middle panel) and studied by confocal immunofluorescence microscopy. The right panels show the merged images, with yellow indicating co-localization. Bar, 5 µm.
Fig. 5. Overexpressed Hrs recruits dynamin and Eps15 to endosomes. BHK cells were transfected with myc-tagged Hrs (A and B) or Hrs1–706 (C) and permeabilized with 0.05% saponin prior to fixation. They were stained with anti-myc (left panels), anti-dynamin (A, middle panel) or anti-Eps15 (B and C, middle panels) and studied by confocal immunofluorescence microscopy. The right panels show the merged images, with yellow indicating co-localization. Bar, 5 µm.
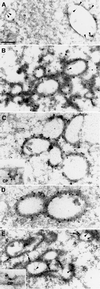
Fig. 6. Electron microscopy of Hrs-overexpressing endosomes. (A) Early endosomes from a non-transfected BHK cell that prior to fixation had endocytosed 5 nm BSA–gold (arrowheads) for 10 min. (B) Localization of Hrs in a myc-Hrs-transfected BHK cell that prior to fixation had endocytosed 5 nm BSA–gold (arrowheads) for 1 h. Thawed cryosections were labelled with anti-myc antibodies followed by rabbit anti-mouse antibodies and 15 nm protein A–gold. Note the coated appearance and clustering of the Hrs-positive compartments. (C) Immunoelectron microscopic localization of clathrin in myc-Hrs-transfected cells. Thawed cryosections were double labelled using rabbit anti-clathrin LC (10 nm gold) followed by mouse anti-myc and rabbit anti-mouse antibodies (15 nm gold). The labelling showed that clathrin localized to the characteristic myc-Hrs-induced endosomal coat. (C, inset) A clathrin-coated pit labelled with goat anti-clathrin followed by rabbit anti-goat and 15 nm protein A–gold. (D) Immunoelectron microscopic localization of Eps15 in myc-Hrs-transfected cells. Thawed cryosections were double labelled with anti-Eps15 (10 nm protein A–gold) followed by labelling for the myc epitope (15 nm protein A–gold). There is a strong Eps15 labelling on the characteristic Hrs-positive endosomal coat. (E) Immunoelectron microscopic localization of dynamin in myc-Hrs-transfected cells. Thawed cryosections of cells that had endocytosed 5 nm BSA–gold for 1 h (arrowheads) were labelled with mouse anti-dynamin 1 antibody (Hudy 1) followed by rabbit anti-mouse antibodies and 15 nm protein A–gold (arrows). In cells showing the characteristic clustering of coated endosomes due to Hrs overexpression, labelling of dynamin is localized to the endosomal coat as well as to typical clathrin-coated pits at the plasma membrane (CP, inset). Bar, 200 nm.
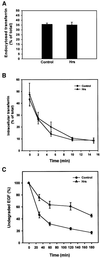
Fig. 7. Overexpressed Hrs does not interfere with transferrin endocytosis or recycling, but inhibits EGF degradation. (A) Effect of Hrs on transferrin endocytosis. BHK cells were transfected with transferrin receptor alone (control) or co-transfected with transferrin receptor and Hrs for 6 h. The cells were incubated with Ru-tag-labelled transferrin for 15 min at 37°C and surface-bound transferrin was removed by pronase. The amount of cell-associated transferrin was measured using an ORIGEN analyser, and endocytosed transferrin is presented as a percentage of total cell-associated transferrin. Error bars represent the SEM of three independent experiments performed in duplicate. (B) Effect of Hrs on transferrin recycling. BHK cells were transfected with transferrin receptor alone (control) or co-transfected with transferrin receptor and Hrs for 6 h. The cells were incubated with Ru-tag-labelled transferrin for 30 min at 37°C and surface-bound transferrin was removed in a portion of the wells by MESNA. The MESNA-treated cells were incubated further for the times indicated. The amount of cell-associated transferrin was measured using an ORIGEN analyser, and intracellular transferrin is presented as a percentage of total cell-associated transferrin. Error bars represent the SEM of two independent experiments performed in duplicate. (C) Effect of Hrs on EGF degradation. HEp-2 cells were transfected with myc-tagged Hrs for 6 h, and rhodamine-EGF was internalized for 1 h at 37°C. The cells were washed and incubated further for different times up to 3 h. The cells were permeabilized with 0.05% saponin prior to fixation. They were stained with anti-myc and studied by confocal immunofluorescence microscopy. The mean intensity of the rhodamine-EGF signal from 10 transfected and 10 non-transfected cells from each time point was quantified and is presented as the percentage of undegraded EGF compared with the rhodamine-EGF signal before chase. Error bars denote± SEM.
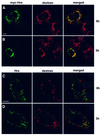
Fig. 8. Overexpressed Hrs inhibits EGF trafficking from early to late endosomes. HEp-2 cells were transfected with myc-tagged Hrs for 6 h and incubated with rhodamine-EGF for 1 h at 37°C (A–F). A portion of the cells was washed and incubated further for 3 h (G–L). The cells were permeabilized with 0.05% saponin prior to fixation. They were stained with anti-myc (A and G) or anti-EEA1 antibodies (C and I) and studied by confocal immunofluorescence microscopy. Turquoise indicates co-localization between Hrs and EEA1 (E and K). Purple indicates co-localization between EGF and EEA1 (F and L). Yellow indicates co-localization between Hrs and EGF (D and J). Bar, 5 µm.
Fig. 9. The transport of endocytosed dextran from early to late endosomes is inhibited in Hrs-overexpressing cells. HEp-2 cells were transfected with myc-tagged Hrs for 6 h and incubated with biotin–dextran for 1 h at 37°C (A). The cells were washed and incubated further for 3 h (B). (C) A non-transfected cell incubated with biotin–dextran for 1 h at 37°C. (D) A non-transfected cell after 3 h of chase. The cells were stained with anti-myc (left panels A and B), anti-Hrs (left panels C and D) or streptavidin–Cy3 (middle panels) and studied by confocal fluorescence microscopy. The right panels show the merged images, with yellow indicating co-localization. Bar, 5 µm.
Fig. 10. Wortmannin disrupts the association of Hrs and clathrin with transferrin-positive endosomes. HEp-2 cells were incubated with Alexa488–transferrin for 30 min at 37°C in the absence (A) or presence (B) of 100 nM wortmannin. The cells were permeabilized with 0.05% saponin prior to fixation in order to remove cytosolic proteins. They were stained with anti-Hrs (left panels) and anti-clathrin (middle panels) and studied by confocal fluorescence microscopy. Arrows indicate examples of co-localization between Hrs, clathrin and internalized transferrin. Arrowheads point at the presumed TGN region stained with anti-clathrin. Bar, 5 µm.
Similar articles
- Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes.
Raiborg C, Bache KG, Gillooly DJ, Madshus IH, Stang E, Stenmark H. Raiborg C, et al. Nat Cell Biol. 2002 May;4(5):394-8. doi: 10.1038/ncb791. Nat Cell Biol. 2002. PMID: 11988743 - Endosomal localization and receptor dynamics determine tyrosine phosphorylation of hepatocyte growth factor-regulated tyrosine kinase substrate.
Urbé S, Mills IG, Stenmark H, Kitamura N, Clague MJ. Urbé S, et al. Mol Cell Biol. 2000 Oct;20(20):7685-92. doi: 10.1128/MCB.20.20.7685-7692.2000. Mol Cell Biol. 2000. PMID: 11003664 Free PMC article. - Flat clathrin coats on endosomes mediate degradative protein sorting by scaffolding Hrs in dynamic microdomains.
Raiborg C, Wesche J, Malerød L, Stenmark H. Raiborg C, et al. J Cell Sci. 2006 Jun 15;119(Pt 12):2414-24. doi: 10.1242/jcs.02978. Epub 2006 May 23. J Cell Sci. 2006. PMID: 16720641 - Function of Hrs in endocytic trafficking and signalling.
Raiborg C, Bache KG, Mehlum A, Stenmark H. Raiborg C, et al. Biochem Soc Trans. 2001 Aug;29(Pt 4):472-5. doi: 10.1042/bst0290472. Biochem Soc Trans. 2001. PMID: 11498011 Review. - Hrs and hbp: possible regulators of endocytosis and exocytosis.
Komada M, Kitamura N. Komada M, et al. Biochem Biophys Res Commun. 2001 Mar;281(5):1065-9. doi: 10.1006/bbrc.2001.4441. Biochem Biophys Res Commun. 2001. PMID: 11243842 Review.
Cited by
- Phagosome maturation: aging gracefully.
Vieira OV, Botelho RJ, Grinstein S. Vieira OV, et al. Biochem J. 2002 Sep 15;366(Pt 3):689-704. doi: 10.1042/BJ20020691. Biochem J. 2002. PMID: 12061891 Free PMC article. Review. - Endosomal dynamics of Met determine signaling output.
Hammond DE, Carter S, McCullough J, Urbé S, Vande Woude G, Clague MJ. Hammond DE, et al. Mol Biol Cell. 2003 Apr;14(4):1346-54. doi: 10.1091/mbc.e02-09-0578. Mol Biol Cell. 2003. PMID: 12686592 Free PMC article. - Exosomal transmission of viruses, a two-edged biological sword.
Mardi N, Haiaty S, Rahbarghazi R, Mobarak H, Milani M, Zarebkohan A, Nouri M. Mardi N, et al. Cell Commun Signal. 2023 Jan 23;21(1):19. doi: 10.1186/s12964-022-01037-5. Cell Commun Signal. 2023. PMID: 36691072 Free PMC article. Review. - O-GlcNAcylation regulates epidermal growth factor receptor intracellular trafficking and signaling.
Wu L, Cheng Y, Geng D, Fan Z, Lin B, Zhu Q, Li J, Qin W, Yi W. Wu L, et al. Proc Natl Acad Sci U S A. 2022 Mar 8;119(10):e2107453119. doi: 10.1073/pnas.2107453119. Epub 2022 Mar 3. Proc Natl Acad Sci U S A. 2022. PMID: 35239437 Free PMC article. - Lysosomal trafficking functions of mucolipin-1 in murine macrophages.
Thompson EG, Schaheen L, Dang H, Fares H. Thompson EG, et al. BMC Cell Biol. 2007 Dec 21;8:54. doi: 10.1186/1471-2121-8-54. BMC Cell Biol. 2007. PMID: 18154673 Free PMC article.
References
- Asao H. et al. (1997) Hrs is associated with STAM, a signal-transducing adaptor molecule. J. Biol. Chem., 272, 32785–32791. - PubMed
- Bean A.J., Davanger,S., Chou,M.F., Gerhardt,B., Tsujimoto,S. and Chang,Y. (2000) Hrs-2 regulates receptor-mediated endocytosis via interactions with Eps15. J. Biol. Chem., 275, 15271–15278. - PubMed
- Burd C.G. and Emr,S.D. (1998) Phosphatidylinositol(3)-phosphate signaling mediated by specific binding to RING FYVE domains. Mol. Cell, 2, 157–162. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources