Localization to the nucleolus is a common feature of coronavirus nucleoproteins, and the protein may disrupt host cell division - PubMed (original) (raw)
Localization to the nucleolus is a common feature of coronavirus nucleoproteins, and the protein may disrupt host cell division
T Wurm et al. J Virol. 2001 Oct.
Abstract
The subcellular localization of transmissible gastroenteritis virus (TGEV) and mouse hepatitis virus (MHV) (group I and group II coronaviruses, respectively) nucleoproteins (N proteins) were examined by confocal microscopy. The proteins were shown to localize either to the cytoplasm alone or to the cytoplasm and a structure in the nucleus. This feature was confirmed to be the nucleolus by using specific antibodies to nucleolin, a major component of the nucleolus, and by confocal microscopy to image sections through a cell expressing N protein. These findings are consistent with our previous report for infectious bronchitis virus (group III coronavirus) (J. A. Hiscox et al., J. Virol. 75:506-512, 2001), indicating that nucleolar localization of the N protein is a common feature of the coronavirus family and is possibly of functional significance. Nucleolar localization signals were identified in the domain III region of the N protein from all three coronavirus groups, and this suggested that transport of N protein to the nucleus might be an active process. In addition, our results suggest that the N protein might function to disrupt cell division. Thus, we observed that approximately 30% of cells transfected with the N protein appeared to be undergoing cell division. The most likely explanation for this is that the N protein induced a cell cycle delay or arrest, most likely in the G(2)/M phase. In a fraction of transfected cells expressing coronavirus N proteins, we observed multinucleate cells and dividing cells with nucleoli (which are only present during interphase). These findings are consistent with the possible inhibition of cytokinesis in these cells.
Figures
FIG. 1
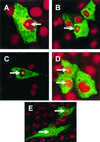
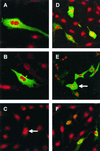
Indirect detection of TGEV (A and B) and MHV (C and D) N proteins in transfected cells and the IBV (E) N protein in transduced cells by immunofluorescence. LLC-PK1 cells (A), L cells (C), and Vero cells (B and D) were transfected with either pCi-TGEV-N or pCi-MHV-N or were transduced with BacIBVN (E). They were incubated for 24 h, fixed, and analyzed by indirect immunofluorescence using appropriate antibodies (green) (see text). Additionally, cells were stained with PI to directly visualize nuclear DNA (red). Differentially fluorescing images were gathered separately from the same 0.5-μm-thick optical section by using a confocal microscope and the appropriate filter. The two images were digitally superimposed to depict the distribution of the appropriate coronavirus N protein and nuclear DNA. Arrow, position of a nucleolus. Magnifications, ×62 (A to D) and ×16 (2.73 zoom) (E).
FIG. 2
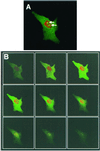
Detection of TGEV N proteins by indirect immunofluorescence in transfected cells. Vero cells were transfected with pCi-TGEV-N, incubated for 24 h, fixed, and analyzed by indirect immunofluorescence using appropriate antibodies (green) (see text). Additionally, cells were stained with PI to visualize nuclear DNA (red). Differentially fluorescing images were gathered separately from the same 0.5-μm-thick optical sections by using a confocal microscope and the appropriate filter. The two images were digitally superimposed to depict the distribution of TGEV N protein and nuclear DNA (A). Arrow, position of a nucleolus. (B) The confocal microscope was used to take 0.1-μm-thick sections of the cell shown in panel A. The section on the top left is nearest the coverslip, and the section on the bottom right is nearest the media. Magnification, ×62 (and ×2 zoom).
FIG. 3
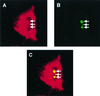
Detection of MHV N protein by indirect immunofluorescence in transfected cells. HeLa cells were transfected with pCi-MHV-N, incubated for 24 h, and analyzed by indirect immunofluorescence using appropriate antibodies to detect the N protein (red) and nucleolin (green) (see text). Differentially fluorescing images were gathered separately from the same 0.5-μm-thick optical section by using a confocal microscope and the appropriate filter. The two images (A and B) were digitally superimposed to depict the distribution of the MHV N protein and nucleolin (C). Yellow indicates colocalization. Magnification, ×61.
FIG. 4
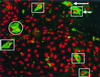
Detection of MHV N proteins by indirect immunofluorescence in transfected cells. Vero cells were transfected with pCi-MHV-N, incubated for 24 h, fixed, and analyzed by indirect immunofluorescence using appropriate antibodies (green) (see text). Additionally, cells were stained with PI to visualize nuclear DNA (red). Differentially fluorescing images were gathered separately from the same 0.5-μm-thick optical sections by using a confocal microscope and the appropriate filter. The two images were digitally superimposed to depict the distribution of the MHV N protein and nuclear DNA. Squares, cells in which the MHV N protein localized to the cytoplasm; circles, cells where the MHV N protein localized to both the cytoplasm and the nucleolus. Magnification, ×16.
FIG. 5
Detection of TGEV N proteins (A, B, and E) and influenza B virus NP (F) by indirect immunofluorescence and detection of EGFP (D) (all green) and cell nuclei (A to F) (red) by direct immunofluorescence. Vero cells were transfected with either pCi-TGEV-N, pCDNA3-NP, or pEGFP, incubated for 24 h, fixed, and analyzed by indirect immunofluorescence using appropriate techniques (see text). Additionally, cells were stained with PI to directly visualize nuclear DNA. Differentially fluorescing images were gathered separately from the same 0.5-μm-thick optical sections by using a confocal microscope and the appropriate filter. The two images were digitally superimposed to depict the distribution of the appropriate protein and nuclear DNA. Magnification, ×62.
Similar articles
- Interaction of the coronavirus nucleoprotein with nucleolar antigens and the host cell.
Chen H, Wurm T, Britton P, Brooks G, Hiscox JA. Chen H, et al. J Virol. 2002 May;76(10):5233-50. doi: 10.1128/jvi.76.10.5233-5250.2002. J Virol. 2002. PMID: 11967337 Free PMC article. - Changes in nucleolar morphology and proteins during infection with the coronavirus infectious bronchitis virus.
Dove BK, You JH, Reed ML, Emmett SR, Brooks G, Hiscox JA. Dove BK, et al. Cell Microbiol. 2006 Jul;8(7):1147-57. doi: 10.1111/j.1462-5822.2006.00698.x. Cell Microbiol. 2006. PMID: 16819967 Free PMC article. - Characterization of Localization and Export Signals of Bovine Torovirus Nucleocapsid Protein Responsible for Extensive Nuclear and Nucleolar Accumulation and Their Importance for Virus Growth.
Ujike M, Kawachi Y, Matsunaga Y, Etho Y, Asanuma H, Kamitani W, Taguchi F. Ujike M, et al. J Virol. 2021 Jan 13;95(3):e02111-20. doi: 10.1128/JVI.02111-20. Print 2021 Jan 13. J Virol. 2021. PMID: 33177195 Free PMC article. - The coronavirus nucleocapsid is a multifunctional protein.
McBride R, van Zyl M, Fielding BC. McBride R, et al. Viruses. 2014 Aug 7;6(8):2991-3018. doi: 10.3390/v6082991. Viruses. 2014. PMID: 25105276 Free PMC article. Review.
Cited by
- Identification of the interaction between vimentin and nucleocapsid protein of transmissible gastroenteritis virus.
Zhang X, Shi H, Chen J, Shi D, Dong H, Feng L. Zhang X, et al. Virus Res. 2015 Mar 16;200:56-63. doi: 10.1016/j.virusres.2014.12.013. Epub 2014 Dec 20. Virus Res. 2015. PMID: 25533531 Free PMC article. - An RNA-Seq analysis of coronavirus in the skin of the Pangolin.
Deng S, Tian X, Belshaw R, Zhou J, Zhang S, Yang Y, Huang C, Chen W, Qiu H, Choo SW. Deng S, et al. Sci Rep. 2024 Jan 9;14(1):910. doi: 10.1038/s41598-024-51261-x. Sci Rep. 2024. PMID: 38195813 Free PMC article. - Identification of cellular proteome using two-dimensional difference gel electrophoresis in ST cells infected with transmissible gastroenteritis coronavirus.
Zhang X, Shi HY, Chen JF, Shi D, Lang HW, Wang ZT, Feng L. Zhang X, et al. Proteome Sci. 2013 Jul 16;11(1):31. doi: 10.1186/1477-5956-11-31. Proteome Sci. 2013. PMID: 23855489 Free PMC article. - Conserved residues in the UL24 protein of herpes simplex virus 1 are important for dispersal of the nucleolar protein nucleolin.
Bertrand L, Leiva-Torres GA, Hyjazie H, Pearson A. Bertrand L, et al. J Virol. 2010 Jan;84(1):109-18. doi: 10.1128/JVI.01428-09. J Virol. 2010. PMID: 19864385 Free PMC article. - The cell cycle and virus infection.
Emmett SR, Dove B, Mahoney L, Wurm T, Hiscox JA. Emmett SR, et al. Methods Mol Biol. 2005;296:197-218. doi: 10.1385/1-59259-857-9:197. Methods Mol Biol. 2005. PMID: 15576934 Free PMC article.
References
- Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson J D. Molecular biology of the cell. 3rd ed. New York, N.Y: Garland Publishing; 1994. pp. 381–382.
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1987.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources