Widespread collaboration of Isw2 and Sin3-Rpd3 chromatin remodeling complexes in transcriptional repression - PubMed (original) (raw)
Widespread collaboration of Isw2 and Sin3-Rpd3 chromatin remodeling complexes in transcriptional repression
T G Fazzio et al. Mol Cell Biol. 2001 Oct.
Abstract
The yeast Isw2 chromatin remodeling complex functions in parallel with the Sin3-Rpd3 histone deacetylase complex to repress early meiotic genes upon recruitment by Ume6p. For many of these genes, the effect of an isw2 mutation is partially masked by a functional Sin3-Rpd3 complex. To identify the full range of genes repressed or activated by these factors and uncover hidden targets of Isw2-dependent regulation, we performed full genome expression analyses using cDNA microarrays. We find that the Isw2 complex functions mainly in repression of transcription in a parallel pathway with the Sin3-Rpd3 complex. In addition to Ume6 target genes, we find that many Ume6-independent genes are derepressed in mutants lacking functional Isw2 and Sin3-Rpd3 complexes. Conversely, we find that ume6 mutants, but not isw2 sin3 or isw2 rpd3 double mutants, have reduced fidelity of mitotic chromosome segregation, suggesting that one or more functions of Ume6p are independent of Sin3-Rpd3 and Isw2 complexes. Chromatin structure analyses of two nonmeiotic genes reveals increased DNase I sensitivity within their regulatory regions in an isw2 mutant, as seen previously for one meiotic locus. These data suggest that the Isw2 complex functions at Ume6-dependent and -independent loci to create DNase I-inaccessible chromatin structure by regulating the positioning or placement of nucleosomes.
Figures
FIG. 1
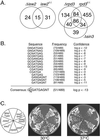
Catalytically inactive and deletion mutations of isw2 and rpd3 have different effects on transcription and growth. (A) The numbers of genes derepressed at least twofold in catalytically inactive (c.i.) and deletion (Δ) mutants of isw2 and rpd3 are compared in Venn diagrams. The number of genes derepressed at least twofold in the Δ_sin3_ mutant is included for comparison. It should be noted that the Venn diagrams tended to overestimate the differences between two mutants, since genes derepressed in both mutants can be derepressed slightly more than twofold in one mutant and slightly less than twofold in the other mutant. Nevertheless, real differences in transcriptional profiles between deletion and catalytically inactive mutants are evident for both isw2 and rpd3. (B) Summary of a regulatory element search of the 489 genes derepressed at least twofold in the rpd3 catalytically inactive mutant but not in the rpd3 deletion mutant. The search parameters allowed for oligonucleotides of eight bases or fewer, permitting degeneracies. Highly significant elements belonging to the same consensus were aligned, and their frequencies and confidence estimates (calculated with the GENESPRING software package) are indicated. (C) Wild-type, deletion, and catalytically inactive mutant yeast cells were streaked on rich (YEPD) plates and incubated at 30 or 37°C. The genotypes of the yeast cells are shown to the left.
FIG. 2
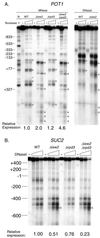
TheIsw2 complex is required for the formation of DNase I-inaccessible chromatin structure at two Ume6-independent loci. (A) Increased DNase I accessibility and improperly positioned nucleosomes at the POT1 locus in isw2 mutant cells. Chromatin structure analysis was performed using micrococcal nuclease (MNase) or DNase I digestion followed by indirect end labeling. MNase and DNase I cleavage sites enhanced in wild-type (WT) or rpd3 mutant cells are marked with circles; those enhanced in isw2 mutant cells are marked with triangles. N, naked DNA control. Relative expression levels indi- cated below each mutant were measured by Northern blotting. For reference, POT1 expression measured by Northern blotting in a ume6 mutant is 1.2-fold that in the wild type. (B) Decreased expression of the SUC2 gene in isw2 mutants despite increased DNase I accessibility of the upstream regulatory region. Chromatin structure analysis was performed using DNase I digestion followed by indirect end labeling. The DNase I cleavage site enhanced in isw2 mutant cells is marked with triangles. Relative expression levels indicated below each mutant were measured by Northern blotting.
Similar articles
- Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters.
Kadosh D, Struhl K. Kadosh D, et al. Cell. 1997 May 2;89(3):365-71. doi: 10.1016/s0092-8674(00)80217-2. Cell. 1997. PMID: 9150136 - The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p.
Goldmark JP, Fazzio TG, Estep PW, Church GM, Tsukiyama T. Goldmark JP, et al. Cell. 2000 Oct 27;103(3):423-33. doi: 10.1016/s0092-8674(00)00134-3. Cell. 2000. PMID: 11081629 - Identification of the Sin3-binding site in Ume6 defines a two-step process for conversion of Ume6 from a transcriptional repressor to an activator in yeast.
Washburn BK, Esposito RE. Washburn BK, et al. Mol Cell Biol. 2001 Mar;21(6):2057-69. doi: 10.1128/MCB.21.6.2057-2069.2001. Mol Cell Biol. 2001. PMID: 11238941 Free PMC article. - Transcriptional regulation of meiosis in budding yeast.
Kassir Y, Adir N, Boger-Nadjar E, Raviv NG, Rubin-Bejerano I, Sagee S, Shenhar G. Kassir Y, et al. Int Rev Cytol. 2003;224:111-71. doi: 10.1016/s0074-7696(05)24004-4. Int Rev Cytol. 2003. PMID: 12722950 Review. - Chromatin remodeling: a marriage between two families?
Pollard KJ, Peterson CL. Pollard KJ, et al. Bioessays. 1998 Sep;20(9):771-80. doi: 10.1002/(SICI)1521-1878(199809)20:9<771::AID-BIES10>3.0.CO;2-V. Bioessays. 1998. PMID: 9819566 Review.
Cited by
- The chromatin remodeling complex NoRC targets HDAC1 to the ribosomal gene promoter and represses RNA polymerase I transcription.
Zhou Y, Santoro R, Grummt I. Zhou Y, et al. EMBO J. 2002 Sep 2;21(17):4632-40. doi: 10.1093/emboj/cdf460. EMBO J. 2002. PMID: 12198165 Free PMC article. - The Torso signaling pathway modulates a dual transcriptional switch to regulate tailless expression.
Chen YC, Lin SI, Chen YK, Chiang CS, Liaw GJ. Chen YC, et al. Nucleic Acids Res. 2009 Mar;37(4):1061-72. doi: 10.1093/nar/gkn1036. Epub 2009 Jan 7. Nucleic Acids Res. 2009. PMID: 19129218 Free PMC article. - Fkh1 and Fkh2 associate with Sir2 to control CLB2 transcription under normal and oxidative stress conditions.
Linke C, Klipp E, Lehrach H, Barberis M, Krobitsch S. Linke C, et al. Front Physiol. 2013 Jul 12;4:173. doi: 10.3389/fphys.2013.00173. eCollection 2013. Front Physiol. 2013. PMID: 23874301 Free PMC article. - Ssn6-Tup1 requires the ISW2 complex to position nucleosomes in Saccharomyces cerevisiae.
Zhang Z, Reese JC. Zhang Z, et al. EMBO J. 2004 Jun 2;23(11):2246-57. doi: 10.1038/sj.emboj.7600227. Epub 2004 Apr 29. EMBO J. 2004. PMID: 15116071 Free PMC article. - ASH1-catalyzed H3K36 methylation drives gene repression and marks H3K27me2/3-competent chromatin.
Bicocca VT, Ormsby T, Adhvaryu KK, Honda S, Selker EU. Bicocca VT, et al. Elife. 2018 Nov 23;7:e41497. doi: 10.7554/eLife.41497. Elife. 2018. PMID: 30468429 Free PMC article.
References
- Alland L, Muhle R, Hou H, Jr, Potes J, Chin L, Schreiber-Agus N, DePinho R A. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. - PubMed
- Bussemaker H J, Li H, Siggia E D. Regulatory element detection using correlation with expression. Nat Genet. 2001;27:167–171. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 CA015704/CA/NCI NIH HHS/United States
- CA74841/CA/NCI NIH HHS/United States
- GM58465/GM/NIGMS NIH HHS/United States
- R29 CA074841/CA/NCI NIH HHS/United States
- R01 GM058465/GM/NIGMS NIH HHS/United States
- R01 CA074841/CA/NCI NIH HHS/United States
- 5P30 CA15704-28/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases