Nuclear entry mechanism of rat PER2 (rPER2): role of rPER2 in nuclear localization of CRY protein - PubMed (original) (raw)
Nuclear entry mechanism of rat PER2 (rPER2): role of rPER2 in nuclear localization of CRY protein
K Miyazaki et al. Mol Cell Biol. 2001 Oct.
Abstract
Mammalian PERIOD2 protein (PER2) is the product of a clock gene that controls circadian rhythms, because PER2-deficient mice have an arrhythmic phenotype. The nuclear entry regulation of clock gene products is a key step in proper circadian rhythm formation in both Drosophila and mammals, because the periodic transcription of clock genes is controlled by an intracellular, oscillating, negative feedback loop. The present study used deletion mutants of rat PER2 (rPER2) to identify the functional nuclear localization signal (NLS) in rPER2. The elimination of putative NLS (residues 778 to 794) from the rPER2 fragment resulted in the loss of nuclear entry activity. Adding the NLS to the cytosolic protein (bacterial alkaline phosphatase) translocates the fusion protein to the nuclei. The data indicate the presence of a functional NLS in rPER2. Furthermore, intact rPER2 was preferentially translocated from the cytoplasm to the nucleus when coexpressed with human CRY1 (hCRY1). However, rPER2 mutants lacking a carboxyl-terminal domain could not enter the nucleus even in the presence of hCRY1. In addition, coexpression of the nuclear localization domain (residues 512 to 794) lacking rPER2 and CRY1 changed the subcellular localization of CRY1 from the nucleus to the cytoplasm. In vitro protein interaction studies demonstrated that the carboxyl-terminal domain of rPER2 is essential for binding to CRY1. The data suggested that both the rPER2 NLS and carboxyl-terminal CRY binding domain are essential for nuclear entry of the rPER2-CRY1 complex.
Figures
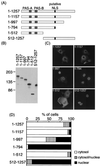
FIG. 1
Subcellular localization of truncated rPER2 mutants in COS-1 cells. (A) Diagrammatic representation of constructs used to identify the rPER2 NLD. PAS-A, PAS-B, and the putative bipartite NLS are shown as shaded and solid boxes. (B) Confirmation of the size of proteins expressed in COS-1 cells determined by immunoblotting. Sizes are indicated in kilodaltons. Anti-rPER2 antiserum was the first antibody. (C) Representative micrographs show the subcellular localization of rPER2. COS-1 cells were transiently transfected with truncated constructs 1–1257, 1–1157, 1–997, 1–794, 1–512, and 512–1257. Forty-eight hours later, cells were fixed and expressed proteins were visualized using anti-rPER2 antiserum and fluorescein isothiocyanate-conjugated secondary antibody. (D) Quantitative analysis of the above. Subcellular localization was categorized as cytoplasm, cytoplasm and nucleus, and nucleus. The ratio of cells with predominant localization to the total transfected cells was determined by counting 50 to 100 cells three to five times in each experiment under light microscopy.
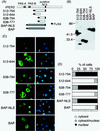
FIG. 2
Expression of NLDs of rPER2 and rPER2 NLS-tagged BAP (NLS-BAP). (A) Schematic diagrams of four constructs covering the NLD of rPER2. All fragments were tagged with FLAG at the amino terminus. rPER2 NLS was inserted at the carboxyl terminus of BAP. (B) The molecular size in kilodaltons of FLAG-tagged NLD fragments and BAP fragments was confirmed by immunoblotting using anti-FLAG M2 monoclonal antibody. (C) Subcellular localization of rPER2 NLD mutants (512–794, 512–644, 638–794, and 638–777) expressed in COS-1 cells and examined by immunofluorescence microscopy. Fragments of rPER2 NLD mutants were stained with a combination of anti-FLAG monoclonal antibody M2 and fluorescein isothiocyanate-conjugated anti-mouse IgG (green, left panels), and nuclei were visualized with DAPI (blue, right panels). The distribution of NLS-BAP and BAP was also confirmed as described above. (D) Quantitative analysis of above as described in the legend for Fig. 1D.
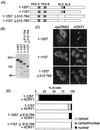
FIG. 3
Role of two rPER2 domains in nuclear localization by CRY1 coexpression. (A) Schematic representation of truncated rPER2 constructs. The position of the rPER2 NLD is shown by shaded boxes as PAS-A and PAS-B. The putative NLS is shown as a solid box. Internal portion of constructs 1–1257 Δ512–794 and 1–1157 Δ512–794, containing NLD, was deleted (amino acids 512 to 794, dotted line). The carboxyl-terminal portion of clones 1–1157 Δ512–794 was also deleted to form clone 1–1157. (B) Molecular size in kilodaltons of clones with a truncation of the NLD was confirmed by immunoblotting using anti-rPER2 polyclonal antibody. (C) Full-length (1–1257) or truncated (1–1157 and 1–1257 Δ512–794) clones with a deletion of rPER2 were cotransfected with hCRY1-pcDNA3.1-His (right panels, + hCRY1) in COS-1 cells. Control used the mock vector, pcDNA3, instead of hCRY1-pcDNA3.1-His (left panels, + pcDNA3). Localization was examined by immunocytochemistry using anti-rPER2 antibody. (D) Quantitation of above as described in the legend for Fig. 1D.
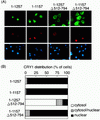
FIG. 4
Subcellular distribution of hCRY1 is affected by cotransfection with rPER2 mutants in COS-1 cells. (A) Full-length (1–1257) and deletion clones truncated at NLDs and/or the carboxyl-terminal portion were coexpressed with Xpress-tagged hCRY1. Transfected cells were triply stained with anti-rPER2 antibodies (green), anti-Xpress antibodies (red), and DAPI (blue). Anti-Xpress antibodies recognized recombinant hCRY and DAPI-stained nuclei. (B) Quantitative analysis of experiment shown in Fig. 4A as described in the legend for Fig. 1D.
FIG. 5
Coimmunoprecipitation shows that hCRY1 interacts with rPER2 carboxyl-terminal domain. (A) Schematic representation of rPER2 mutant constructs for coimmunoprecipitation. The positions of rPER2 NLD, PAS-A, and PAS-B are shown by shaded boxes. The rPRE2 NLS is shown as a solid box. The mutants with a deletion at the amino-terminal half (512–1257 and 512–1157) and at the carboxyl-terminal fragment of rPER2 (1157–1257) were tagged with FLAG at their amino-terminal ends. (B) Total lysates from cells coexpressing hCRY1 and truncation mutants of rPER2 were immunoprecipitated (IP) and blotted with anti-rPER2 antibodies (top panel) and were detected using anti-Xpress antibodies (lower panel). Results were similar in replicate experiments. Arrow indicates position of hCRY1. (C) Identification of the CRY1 binding domain of rPER2. The FLAG-tagged truncation mutants of rPER2 proteins (1157–1257, 512–1257, and 512–1157) were expressed with hCRY1. Control was vector pcDNA3 or pFLAG instead of hCRY1 or rPER2 mutants. Lysates from transfected cells were immunoprecipitated (IP) and blotted with anti-FLAG antibodies (top panel) and were visualized using anti-Xpress antibodies (lower panel). The arrow indicates the position of hCRY1. Asterisks indicate nonspecific band corresponding to Ig derived from M2 monoclonal antibodies. Numerals indicate molecular weights in thousands.
Similar articles
- PER2 controls circadian periods through nuclear localization in the suprachiasmatic nucleus.
Miyazaki K, Wakabayashi M, Chikahisa S, Sei H, Ishida N. Miyazaki K, et al. Genes Cells. 2007 Nov;12(11):1225-34. doi: 10.1111/j.1365-2443.2007.01129.x. Genes Cells. 2007. PMID: 17986006 - Dimerization and nuclear entry of mPER proteins in mammalian cells.
Yagita K, Yamaguchi S, Tamanini F, van Der Horst GT, Hoeijmakers JH, Yasui A, Loros JJ, Dunlap JC, Okamura H. Yagita K, et al. Genes Dev. 2000 Jun 1;14(11):1353-63. Genes Dev. 2000. PMID: 10837028 Free PMC article. - Interactivating feedback loops within the mammalian clock: BMAL1 is negatively autoregulated and upregulated by CRY1, CRY2, and PER2.
Yu W, Nomura M, Ikeda M. Yu W, et al. Biochem Biophys Res Commun. 2002 Jan 25;290(3):933-41. doi: 10.1006/bbrc.2001.6300. Biochem Biophys Res Commun. 2002. PMID: 11798163 - [Molecular mechanisms of biological clock: from molecular rhythms to physiological rhythms].
Okamura H. Okamura H. No To Shinkei. 2003 Jan;55(1):5-11. No To Shinkei. 2003. PMID: 12649895 Review. Japanese. No abstract available. - Cellular and molecular basis of circadian timing in mammals.
Reppert SM. Reppert SM. Semin Perinatol. 2000 Aug;24(4):243-6. doi: 10.1053/sper.2000.9122. Semin Perinatol. 2000. PMID: 10975430 Review.
Cited by
- Transcriptional activity and nuclear localization of Cabut, the Drosophila ortholog of vertebrate TGF-β-inducible early-response gene (TIEG) proteins.
Belacortu Y, Weiss R, Kadener S, Paricio N. Belacortu Y, et al. PLoS One. 2012;7(2):e32004. doi: 10.1371/journal.pone.0032004. Epub 2012 Feb 16. PLoS One. 2012. PMID: 22359651 Free PMC article. - mPER1-mediated nuclear export of mCRY1/2 is an important element in establishing circadian rhythm.
Loop S, Katzer M, Pieler T. Loop S, et al. EMBO Rep. 2005 Apr;6(4):341-7. doi: 10.1038/sj.embor.7400372. EMBO Rep. 2005. PMID: 15791269 Free PMC article. - Nucleocytoplasmic shuttling and mCRY-dependent inhibition of ubiquitylation of the mPER2 clock protein.
Yagita K, Tamanini F, Yasuda M, Hoeijmakers JH, van der Horst GT, Okamura H. Yagita K, et al. EMBO J. 2002 Mar 15;21(6):1301-14. doi: 10.1093/emboj/21.6.1301. EMBO J. 2002. PMID: 11889036 Free PMC article. - Genetic insights on sleep schedules: this time, it's PERsonal.
Chong SY, Ptáček LJ, Fu YH. Chong SY, et al. Trends Genet. 2012 Dec;28(12):598-605. doi: 10.1016/j.tig.2012.08.002. Epub 2012 Aug 28. Trends Genet. 2012. PMID: 22939700 Free PMC article. Review. - Phosphorylation of clock protein PER1 regulates its circadian degradation in normal human fibroblasts.
Miyazaki K, Nagase T, Mesaki M, Narukawa J, Ohara O, Ishida N. Miyazaki K, et al. Biochem J. 2004 May 15;380(Pt 1):95-103. doi: 10.1042/BJ20031308. Biochem J. 2004. PMID: 14750904 Free PMC article.
References
- Albrecht U, Sun Z S, Eichele G, Lee C C. A differential response of two putative mammalian circadian regulators, mper1 and mper2, to light. Cell. 1997;91:1055–1064. - PubMed
- Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. - PubMed
- Dunlap J C. Molecular bases for circadian clocks. Cell. 1999;96:271–290. - PubMed
- Field M D, Maywood E S, O'Brien J A, Weaver D R, Reppert S M, Hastings M H. Analysis of clock proteins in mouse SCN demonstrates phylogenetic divergence of the circadian clockwork and resetting mechanisms. Neuron. 2000;25:437–447. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases