Argus--a new database system for Web-based analysis of multiple microarray data sets - PubMed (original) (raw)
Argus--a new database system for Web-based analysis of multiple microarray data sets
J Comander et al. Genome Res. 2001 Sep.
Abstract
The ongoing revolution in microarray technology allows biologists studying gene expression to routinely collect >10(5) data points in a given experiment. Widely accessible and versatile database software is required to process this large amount of raw data into a format that facilitates the development of new biological insights. Here, we present a novel microarray database software system, named Argus, designed to process, analyze, manage, and publish microarray data. Argus imports the intensities and images of externally quantified microarray spots, performs normalization, and calculates ratios of gene expression between conditions. The database can be queried locally or over the Web, providing a convenient format for Web-publishing entire microarray data sets. Searches for regulated genes can be conducted across multiple experiments, and the integrated results incorporate images of the actual hybridization spots for artifact screening. Query results are presented in a clone- or gene-oriented fashion to rapidly identify highly regulated genes, and scatterplots of expression ratios allow an individual ratio to be interpreted in the context of all data points in the experiment. Algorithms were developed to optimize response times for queries of regulated genes. Supporting databases are updated easily to maintain current gene identity information, and hyperlinks to the Web provide access to descriptions of gene function. Query results also can be exported for higher-order analyses of expression patterns. This combination of features currently is not available in similar software. Argus is available at http://vessels.bwh.harvard.edu/software/Argus.
Figures
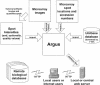
Figure 1
Data flow diagram. First, microarray spots are quantified using an external program (arrow, top left).
Argus
then imports all data necessary to analyze a set of microarray experiments (counterclockwise from right): the latest version of the UniGene database, a description of the location of each clone on the arrays, the actual images of the scanned microarrays, and the intensity and quality value of each spot on the arrays.
Argus
processes these data and produces a database and a set of supporting files that are transferred (arrow, bottom right) to a local or centrally managed Web server. Users (bottom center), whether local or at a remote location, can access all analysis features of the interactive database using a Web browser and also can conveniently access remote biological databases (bottom left) for additional gene information.
Figure 2
A typical query for regulated genes. (A) The search form is configured to retrieve clones that were down-regulated at least twofold when comparing condition B to condition A and that have a minimum intensity of 3000, from any array or experiment in the database. (B) Three clones meet these criteria for at least one of their replicate measurements. If desired, ratios from all conditions can be shown on this page. Clicking on the accession number next to fibronectin 1 retrieves all data from that clone (C), including thumbnail images of the actual hybridization spots. Clicking on the image of a spot produces a scatterplot (D) highlighting that data point, in this case showing that the point is outside the scatter of points around the unity line.
Figure 3
Display of multiple clones from the same gene. From the details page of any clone, a new search can be initiated that displays all clones with the same gene name. In both the top and bottom clones, fibroblast growth factor receptor 3 was not regulated in condition B but was down-regulated in conditions C and D. The top clone was nearly five times as intense as the bottom clone, as seen in the Ref Intensity column, yet the intensity ratios between conditions are very similar.
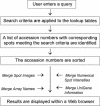
Figure 4
Query processing using lookup tables. When a user submits a query from a Web browser, precalculated lookup tables are used to identify a list of accession numbers that match the search criteria. These accession numbers are sorted and merged with the additional information shown in italics, and the results are displayed in a Web browser window.
Similar articles
- A web based resource characterizing the zebrafish developmental profile of over 16,000 transcripts.
Ouyang M, Garnett AT, Han TM, Hama K, Lee A, Deng Y, Lee N, Liu HY, Amacher SL, Farber SA, Ho SY. Ouyang M, et al. Gene Expr Patterns. 2008 Feb;8(3):171-80. doi: 10.1016/j.gep.2007.10.011. Epub 2007 Nov 7. Gene Expr Patterns. 2008. PMID: 18068546 Free PMC article. - Gene Aging Nexus: a web database and data mining platform for microarray data on aging.
Pan F, Chiu CH, Pulapura S, Mehan MR, Nunez-Iglesias J, Zhang K, Kamath K, Waterman MS, Finch CE, Zhou XJ. Pan F, et al. Nucleic Acids Res. 2007 Jan;35(Database issue):D756-9. doi: 10.1093/nar/gkl798. Epub 2006 Nov 7. Nucleic Acids Res. 2007. PMID: 17090592 Free PMC article. - MILANO--custom annotation of microarray results using automatic literature searches.
Rubinstein R, Simon I. Rubinstein R, et al. BMC Bioinformatics. 2005 Jan 20;6:12. doi: 10.1186/1471-2105-6-12. BMC Bioinformatics. 2005. PMID: 15661078 Free PMC article. - ProbeMatchDB--a web database for finding equivalent probes across microarray platforms and species.
Wang P, Ding F, Chiang H, Thompson RC, Watson SJ, Meng F. Wang P, et al. Bioinformatics. 2002 Mar;18(3):488-9. doi: 10.1093/bioinformatics/18.3.488. Bioinformatics. 2002. PMID: 11934751 - Goober: a fully integrated and user-friendly microarray data management and analysis solution for core labs and bench biologists.
Luo W, Gudipati M, Jung K, Chen M, Marschke KB. Luo W, et al. J Integr Bioinform. 2009 Aug 23;6(1):108. doi: 10.2390/biecoll-jib-2009-108. J Integr Bioinform. 2009. PMID: 20134074
Cited by
- KLF2 Is a novel transcriptional regulator of endothelial proinflammatory activation.
SenBanerjee S, Lin Z, Atkins GB, Greif DM, Rao RM, Kumar A, Feinberg MW, Chen Z, Simon DI, Luscinskas FW, Michel TM, Gimbrone MA Jr, García-Cardeña G, Jain MK. SenBanerjee S, et al. J Exp Med. 2004 May 17;199(10):1305-15. doi: 10.1084/jem.20031132. Epub 2004 May 10. J Exp Med. 2004. PMID: 15136591 Free PMC article. - Vascular endothelium, hemodynamics, and the pathobiology of atherosclerosis.
Gimbrone MA Jr, García-Cardeña G. Gimbrone MA Jr, et al. Cardiovasc Pathol. 2013 Jan-Feb;22(1):9-15. doi: 10.1016/j.carpath.2012.06.006. Epub 2012 Jul 18. Cardiovasc Pathol. 2013. PMID: 22818581 Free PMC article. Review. - ArrayQuest: a web resource for the analysis of DNA microarray data.
Argraves GL, Jani S, Barth JL, Argraves WS. Argraves GL, et al. BMC Bioinformatics. 2005 Dec 1;6:287. doi: 10.1186/1471-2105-6-287. BMC Bioinformatics. 2005. PMID: 16321157 Free PMC article. - Improving the statistical detection of regulated genes from microarray data using intensity-based variance estimation.
Comander J, Natarajan S, Gimbrone MA Jr, García-Cardeña G. Comander J, et al. BMC Genomics. 2004 Feb 27;5(1):17. doi: 10.1186/1471-2164-5-17. BMC Genomics. 2004. PMID: 15113402 Free PMC article.
References
- Aach J, Rindone W, Church GM. Systematic management and analysis of yeast gene expression data. Genome Res. 2000;10:431–445. - PubMed
- Aach J, Bulyk ML, Church GM, Comander J, Derti A, Shendure J. Computational comparison of two draft sequences of the human genome. Nature. 2001;409:856–859. - PubMed
- Boguski MS, Schuler GD. ESTablishing a human transcript map. Nat Genet. 1995;10:369–371. - PubMed
- Bowtell DD. Options available—from start to finish—for obtaining expression data by microarray. Nat Genet. 1999;21:25–32. - PubMed
Publication types
MeSH terms
Grants and funding
- R01LM06538-01/LM/NLM NIH HHS/United States
- T15 LM007092/LM/NLM NIH HHS/United States
- P50-HL56985/HL/NHLBI NIH HHS/United States
- T32 GM007753/GM/NIGMS NIH HHS/United States
- R37-HL511509/HL/NHLBI NIH HHS/United States
- T15LM07092-07/LM/NLM NIH HHS/United States
- T32-HLI07623/HL/NHLBI NIH HHS/United States
- P50 HL056985/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources