Dad1p, third component of the Duo1p/Dam1p complex involved in kinetochore function and mitotic spindle integrity - PubMed (original) (raw)
Dad1p, third component of the Duo1p/Dam1p complex involved in kinetochore function and mitotic spindle integrity
M Enquist-Newman et al. Mol Biol Cell. 2001 Sep.
Free PMC article
Abstract
We showed recently that a complex between Duo1p and Dam1p is required for both spindle integrity and kinetochore function in the budding yeast Saccharomyces cerevisiae. To extend our understanding of the functions and interactions of the Duo1p/Dam1p complex, we analyzed the novel gene product Dad1p (for Duo1 and Dam1 interacting). Dad1p physically associates with Duo1p by two-hybrid analysis, coimmunoprecipitates with Duo1p and Dam1p out of yeast protein extracts, and shows interdependent localization with Duo1p and Dam1p to the mitotic spindle. These results indicate that Dad1p functions as a component of the Duo1p/Dam1p complex. Like Duo1p and Dam1p, Dad1p also localizes to kinetochore regions in chromosomes spreads. Here, we also demonstrate by chromatin immunoprecipitation that Duo1p, Dam1p, and Dad1p associate specifically with centromeric DNA in a manner that is dependent upon Ndc10 and partially dependent upon the presence of microtubules. To explore the functions of Dad1p in vivo, we generated a temperature-sensitive allele, dad1-1. This allele shows spindle defects and a mitotic arrest phenotype that is dependent upon the spindle assembly checkpoint. In addition, dad1-1 mutants undergo chromosome mis-segregation at the restrictive temperature, resulting in a dramatic decrease in viability.
Figures
Figure 1
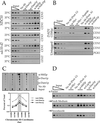
Dad1p has homologs in other fungal species. Residues identical in at least three of the four proteins are boxed and conserved residues are highlighted. Residues mutated in_dad1-1_ are indicated with an asterisk. In pairwise alignments, S. cerevisia_e Dad1p is 29% identical (27 of 94) and 49% conserved (46 of 94) with the A. nidulans homolog (from 13d09a1.r1), 24% identical (22 of 91) and 46% conserved (42 of 91) with the C. albicans homolog (from sequencing of Contig6-2505), and 28% identical (24 of 85) and 52% conserved (44 of 85) with the S. pombe homolog (SPAC16A10.05C). Not shown is a homolog in a closely related_Saccharomycetales species, which is 50% identical (47 of 94) and 68% conserved (64 of 94) with S. cerevisiae Dad1p.
Figure 2
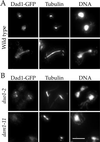
Dad1p localizes to the mitotic spindle in a Duo1p- and Dam1p-dependent manner. (A) Dad1-GFP localization in wild-type cells. Cells were grown to log phase and processed for immunofluorescence staining of Dad1-GFP (anti-GFP antibodies) and tubulin (anti-tubulin antibody), and for DNA (4′,6-diamidino-2-phenylindole [DAPI]). (B) Dad1-GFP localization in_duo1_ and dam1 mutants. Cells were grown at 25°C, shifted to 37°C for 1.5 h, and processed for immunofluorescence as described above. Bar, 5 μm.
Figure 3
Dad1p associates with Duo1p and Dam1p in yeast protein extracts. Protein extracts from wild-type cells and protein extracts from a strain containing HA-tagged Dad1p were incubated with antibodies recognizing Duo1p, Dam1p, HA, or with preimmune serum bound to protein A beads (see MATERIALS AND METHODS). Immunoprecipitated samples were run on a 15% polyacrylamide gel and probed with antibodies recognizing either Duo1p, Dam1p, or HA. Duo1p, Dam1p, and Dad1-HA are specifically immunoprecipiated when antibodies against either Duo1p or Dam1p are used, but not with preimmune serum or in a strain that lacks the HA-tagged Dad1p.
Figure 4
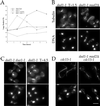
dad1-1 shows mitotic defects. (A) Morphological arrest. Cells were grown to log phase at 25°C and shifted to 37°C at t = 0. The percentage of cells showing a large budded morphology was determined by counting a fixed and sonicated sample. (B) dad1-1 mutant phenotype. Cells were grown at 25°C, shifted to 37°C for 1.5 h, and processed for tubulin immunofluorescence (anti-tubulin antibody) and DNA staining (DAPI). (C) Broken down spindle phenotypes in dad1-1 duo1-1 double mutants or dad1-1 mutants at later time points. Cells were grown at 25°C, shifted to 37°C for 1.5 h or 4.5 h as indicated, and processed for immunofluorescence as described above. (D) Late mitotic mutant phenotypes. Cells were arrested with alpha factor at 25°C, released to fresh prewarmed medium at 37°C for 3 h, and processed for immunofluorescence as described above. Bar, 5 μm.
Figure 5
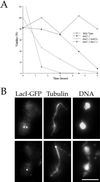
dad1-1 mutants show chromosome segregation defects. (A) Viability of dad1-1 mutants. Cells were grown to log phase at 25°C then shifted to 37°C at t = 0. For each time point, cells were plated onto plates containing YPD medium at 25°C and a sample was counted with the use of a hemacytometer to determine the cells per milliliter of culture. (B) Chromosome mis-segregation observed with the use of LacI-GFP system of chromosome tagging. dad1-1 mad2Δ double mutant cells were grown to log phase at 25°C, shifted to 37°C for 3 h, and processed for immunofluorescence staining for LacI-GFP (anti-GFP antibodies), tubulin (anti-tubulin antibody), and DNA (DAPI). Bar, 5 μm.
Figure 6
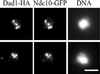
Dad1-HA localizes to kinetochores in spreads of mitotic chromosomes. Cells coexpressing Dad1-HA and Ndc10-GFP fusion proteins were prepared for chromosome spreads as described (Loidl et al., 1998; Cheeseman et al., 2001). They were then processed for immunofluorescence and stained with anti-GFP antibodies to localize Ndc10-GFP and anti-HA antibodies to localize Dad1-HA. Bar, 2.5 μm.
Figure 7
Duo1p, Dam1p, and Dad1p are present at centromeric loci in vivo. (A) Coimmunoprecipitation of CEN DNA with Duo1p and Dam1p. Wild-type (YPH499) and ndc10-42 mutant (PM1021-20A) strains were grown at 23°C in YPD until mid-logarithmic phase (OD600 of ∼0.5), at which point a portion of each culture was transferred to 37°C for 3 h. Formaldehyde cross-linked chromatin prepared from each culture was mock-treated (no antibody) or immunoprecipitated with the indicated antisera. Aliquots of total input material (∼1.0, 0.5, and 0.25 μl of chromatin solution) and coimmunoprecipitated DNA (∼100 μl of chromatin solution for Duo1p and Dam1p or ∼50 μl for Mif2p and Ndc10p) were analyzed by PCR with primers specific for the indicated loci. Yields as a percentage of total input material are indicated in Table 2. Interaction of Duo1p and Dam1p with CEN DNA is centromere-specific and _NDC10_-dependent. (B) Coimmunoprecipitation of CEN DNA with Dad1-13myc. Dad1-13myc (DDY2166) and untagged strains (YPH499) were used for chromatin immunoprecipitation as in A, except an FA lysis buffer was used. To determine the specificity of this immunoprecipitation, myc peptide was in 200 or 500 M excess to myc antibody. (C) Interactions of Duo1p and Dam1p are limited to the CEN DNA. Total input material (∼0.5 μl of chromatin solution) and coimmunoprecipitated DNA (∼100 μl of chromatin solution for Duo1p and Dam1p or ∼50 μl for Mif2p) from YPH499 were analyzed by PCR with the use of primers specific for a series of overlapping_CEN3_-flanking fragments. Below, the percentage of total input material thatcoimmunoprecipitated with each protein is plotted against the center coordinate for each interval amplified. The gray bar indicates the interval that encompasses CEN3. As for Mif2p (and for most centromere components), the interaction of Duo1p and Dam1p with the CEN3 region occurs predominately at CEN3, whereas_CEN3_-flanking regions are enriched to a lesser extent. Identical results were also obtained for Dad1-13myc (our unpublished results). (D) Interaction of Duo1p, Dam1p, and Dad1-myc is partially dependent on microtubules. Dad1-13myc (DDY2166) and untagged (YPH499) strains were arrested in G1 with 5 μM alpha factor for 2 h at 30°C (α factor). Portions of these cells were then washed two times and released into fresh medium (⇒ fresh medium), or medium containing 15 μg/ml nocodazole (⇒ nocodazole) for 1.5 h at 30°C. Microtubule depolyermization was confirmed by indirect immunofluorescence with an anti-β-tubulin antibody. All samples were then processed for chromatin immunoprecipitation as in A, although the FA lysis buffer was used for the Dad1-tagged strain. The data shown corresponds to CEN3 for the Dad1-tagged strain. Yields of both CEN3 and CEN1 as a percentage of total input material were averaged for the tagged and untagged strains and are indicated in Table 3.
Similar articles
- The composition, functions, and regulation of the budding yeast kinetochore.
Biggins S. Biggins S. Genetics. 2013 Aug;194(4):817-46. doi: 10.1534/genetics.112.145276. Genetics. 2013. PMID: 23908374 Free PMC article. Review. - Mitotic spindle integrity and kinetochore function linked by the Duo1p/Dam1p complex.
Cheeseman IM, Enquist-Newman M, Müller-Reichert T, Drubin DG, Barnes G. Cheeseman IM, et al. J Cell Biol. 2001 Jan 8;152(1):197-212. doi: 10.1083/jcb.152.1.197. J Cell Biol. 2001. PMID: 11149931 Free PMC article. - Saccharomyces cerevisiae Duo1p and Dam1p, novel proteins involved in mitotic spindle function.
Hofmann C, Cheeseman IM, Goode BL, McDonald KL, Barnes G, Drubin DG. Hofmann C, et al. J Cell Biol. 1998 Nov 16;143(4):1029-40. doi: 10.1083/jcb.143.4.1029. J Cell Biol. 1998. PMID: 9817759 Free PMC article. - Yeast Dam1p has a role at the kinetochore in assembly of the mitotic spindle.
Jones MH, He X, Giddings TH, Winey M. Jones MH, et al. Proc Natl Acad Sci U S A. 2001 Nov 20;98(24):13675-80. doi: 10.1073/pnas.241417098. Epub 2001 Nov 6. Proc Natl Acad Sci U S A. 2001. PMID: 11698664 Free PMC article. - Dam1 is the right one: phosphoregulation of kinetochore biorientation.
Courtwright AM, He X. Courtwright AM, et al. Dev Cell. 2002 Nov;3(5):610-1. doi: 10.1016/s1534-5807(02)00332-5. Dev Cell. 2002. PMID: 12431367 Review.
Cited by
- Implication of a novel multiprotein Dam1p complex in outer kinetochore function.
Cheeseman IM, Brew C, Wolyniak M, Desai A, Anderson S, Muster N, Yates JR, Huffaker TC, Drubin DG, Barnes G. Cheeseman IM, et al. J Cell Biol. 2001 Dec 24;155(7):1137-45. doi: 10.1083/jcb.200109063. Epub 2001 Dec 24. J Cell Biol. 2001. PMID: 11756468 Free PMC article. - Structural view of the yeast Dam1 complex, a ring-shaped molecular coupler for the dynamic microtubule end.
Wu S, Grishchuk EL. Wu S, et al. Essays Biochem. 2020 Sep 4;64(2):359-370. doi: 10.1042/EBC20190079. Essays Biochem. 2020. PMID: 32579171 Free PMC article. Review. - The composition, functions, and regulation of the budding yeast kinetochore.
Biggins S. Biggins S. Genetics. 2013 Aug;194(4):817-46. doi: 10.1534/genetics.112.145276. Genetics. 2013. PMID: 23908374 Free PMC article. Review. - De novo kinetochore assembly requires the centromeric histone H3 variant.
Collins KA, Castillo AR, Tatsutani SY, Biggins S. Collins KA, et al. Mol Biol Cell. 2005 Dec;16(12):5649-60. doi: 10.1091/mbc.e05-08-0771. Epub 2005 Oct 5. Mol Biol Cell. 2005. PMID: 16207811 Free PMC article. - The DASH complex and Klp5/Klp6 kinesin coordinate bipolar chromosome attachment in fission yeast.
Sanchez-Perez I, Renwick SJ, Crawley K, Karig I, Buck V, Meadows JC, Franco-Sanchez A, Fleig U, Toda T, Millar JB. Sanchez-Perez I, et al. EMBO J. 2005 Aug 17;24(16):2931-43. doi: 10.1038/sj.emboj.7600761. Epub 2005 Aug 4. EMBO J. 2005. PMID: 16079915 Free PMC article.
References
- Ayscough KR, Drubin DG. Immunofluorescence microscopy of yeast cells. In: Celis J, editor. Cell Biology: A Laboratory Handbook. Vol. 2. New York: Academic Press; 1998. pp. 477–485.
- Belmont AS, Straight AF. In vivo visualization of chromosomes using lac operator-repressor binding. Trends Cell Biol. 1998;8:121–124. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM047842/GM/NIGMS NIH HHS/United States
- R01 GM060464/GM/NIGMS NIH HHS/United States
- GM-47842/GM/NIGMS NIH HHS/United States
- R01 GM-60464-02/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases