Introduction of human apolipoprotein E4 "domain interaction" into mouse apolipoprotein E - PubMed (original) (raw)
Introduction of human apolipoprotein E4 "domain interaction" into mouse apolipoprotein E
R L Raffai et al. Proc Natl Acad Sci U S A. 2001.
Abstract
Human apolipoprotein E4 (apoE4) binds preferentially to lower density lipoproteins, including very low density lipoproteins, and is associated with increased risk of atherosclerosis and neurodegenerative disorders, including Alzheimer's disease. This binding preference is the result of the presence of Arg-112, which causes Arg-61 in the amino-terminal domain to interact with Glu-255 in the carboxyl-terminal domain. ApoE2 and apoE3, which have Cys-112, bind preferentially to high density lipoproteins (HDL) and do not display apoE4 domain interaction. Mouse apoE, like apoE4, contains the equivalent of Arg-112 and Glu-255, but lacks the critical Arg-61 equivalent (it contains Thr-61). Thus, mouse apoE does not display apoE4 domain interaction and, as a result, behaves like human apoE3, including preferential binding to HDL. To assess the potential role of apoE4 domain interaction in atherosclerosis and neurodegeneration, we sought to introduce apoE4 domain interaction into mouse apoE. Replacing Thr-61 in mouse apoE with arginine converted the binding preference from HDL to very low density lipoproteins in vitro, suggesting that apoE4 domain interaction could be introduced into mouse apoE in vivo. Using gene targeting in embryonic stem cells, we created mice expressing Arg-61 apoE. Heterozygous Arg-61/wild-type apoE mice displayed two phenotypes found in human apoE4/E3 heterozygotes: preferential binding to lower density lipoproteins and reduced abundance of Arg-61 apoE in the plasma, reflecting its more rapid catabolism. These findings demonstrate the successful introduction of apoE4 domain interaction into mouse apoE in vivo. The Arg-61 apoE mouse model will allow the effects of apoE4 domain interaction in lipoprotein metabolism, atherosclerosis, and neurodegeneration to be determined.
Figures
Figure 1
Generation and characterization of an Arg-61 Apoe allele. (A) The sequence-replacement, gene-targeting strategy. A 7-kb Eco_RI-BstB_1 fragment containing exons 1, 2, and 3 and 5′ flanking sequences was inserted upstream of a neo cassette flanked by_loxP sites. A 1.3-kb fragment containing exon 4 and 3′ flanking sequences was amplified by PCR and cloned into the_Asc_1 site downstream of the neo cassette. Homologous recombination between the gene-targeting vector and the_Apoe locus in ES cells introduced a G-for-C change in the second position of codon 61 at the 3′ end of exon 3 and introduced a unique _Dde_I restriction site. A neo cassette flanked by loxP sites was placed in close proximity to the novel Arg-61 codon in intron 3 to monitor the correct integration of the point mutation. Targeted ES cell clones and mice were identified by digestion of genomic DNA with Eco_RI and Southern blotting with an Apoe exon 4 probe, which revealed an expanded 10.3-kb fragment vs. a wt 8.3-kb fragment. Cre-mediated excision of the neo cassette produces a mutant Apoe allele (designated Arg-61_Apoe).
Figure 2
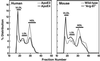
Plasma lipoprotein-binding preferences of human apoE and mouse apoE. Recombinant mouse apoE and human apoE isoforms were radiolabeled, incubated with human plasma, and fractionated into various lipoprotein classes by size-exclusion FPLC.
Figure 3
Comparison of apoE mRNA levels in various tissues and organs from wt (+) mice and Arg-61-targeted mice crossed with Cre-deleter mice (R). (A) Total RNA from various tissues and organs was isolated from wt and Agr-61 mice and subjected to Northern blot analysis with an_Apoe_ exon 4 probe. Tissues and organs included: B, brain; Li, liver; Lu, lung; M, muscle; Si, small intestine; T, testis; H, heart; St, stomach; K, kidney; Sk, skin; Sp, spleen; and E, eye. (B) A control mouse β-actin hybridization is shown.
Figure 4
IEF and Western blot of mouse plasma and CSF apoE. Mouse apoE isoforms in plasma and CSF were monitored by IEF and Western blot detection. Mice were fasted for 4 h before bleeding.
Figure 5
IEF and Western blot of apoE secreted from primary hepatocytes into the culture medium. Primary hepatocytes were isolated from wt and Arg-61/wt heterozygous mice and placed into culture for 2 days, and the medium was collected. Mouse wt and Arg-61 apoE in medium were assessed by IEF and Western blot detection.
Figure 6
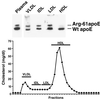
Distribution of wt and Arg-61 apoE in plasma from a heterozygous, gene-targeted mouse on a high-cholesterol diet. Arg-61/wt apoE heterozygous mice were fed the Paigen diet for 6 days to increase VLDL levels. Nonfasted plasma was fractionated by size-exclusion FPLC. (Upper) Fractions corresponding to the different lipoprotein classes were resolved by IEF, and apoE was detected by Western blotting.
Similar articles
- Effect of domain interaction on apolipoprotein E levels in mouse brain.
Ramaswamy G, Xu Q, Huang Y, Weisgraber KH. Ramaswamy G, et al. J Neurosci. 2005 Nov 16;25(46):10658-63. doi: 10.1523/JNEUROSCI.1922-05.2005. J Neurosci. 2005. PMID: 16291938 Free PMC article. - Plasma ApoE4 Levels Are Lower than ApoE2 and ApoE3 Levels, and Not Associated with Plasma Aβ40/42 Ratio as a Biomarker of Amyloid-β Amyloidosis in Alzheimer's Disease.
Nakamura T, Kawarabayashi T, Ueda T, Shimomura S, Hoshino M, Itoh K, Ihara K, Nakaji S, Takatama M, Ikeda Y, Shoji M. Nakamura T, et al. J Alzheimers Dis. 2023;93(1):333-348. doi: 10.3233/JAD-220996. J Alzheimers Dis. 2023. PMID: 36970894 - Apolipoprotein E in lipoprotein metabolism, health and cardiovascular disease.
Marais AD. Marais AD. Pathology. 2019 Feb;51(2):165-176. doi: 10.1016/j.pathol.2018.11.002. Epub 2018 Dec 28. Pathology. 2019. PMID: 30598326 Review. - RANK-Fc: a therapeutic antagonist for RANK-L in myeloma.
Sordillo EM, Pearse RN. Sordillo EM, et al. Cancer. 2003 Feb 1;97(3 Suppl):802-12. doi: 10.1002/cncr.11134. Cancer. 2003. PMID: 12548579 Review.
Cited by
- The influence of 17q21.31 and APOE genetic ancestry on neurodegenerative disease risk.
Harerimana NV, Goate AM, Bowles KR. Harerimana NV, et al. Front Aging Neurosci. 2022 Oct 20;14:1021918. doi: 10.3389/fnagi.2022.1021918. eCollection 2022. Front Aging Neurosci. 2022. PMID: 36337698 Free PMC article. Review. - ApoE isoform-specific differences in behavior and cognition associated with subchronic MPTP exposure.
Torres ERS, Weber Boutros S, Meshul CK, Raber J. Torres ERS, et al. Learn Mem. 2020 Aug 17;27(9):372-379. doi: 10.1101/lm.052126.120. Print 2020 Sep. Learn Mem. 2020. PMID: 32817303 Free PMC article. - A multi-hit hypothesis for an APOE4-dependent pathophysiological state.
Steele OG, Stuart AC, Minkley L, Shaw K, Bonnar O, Anderle S, Penn AC, Rusted J, Serpell L, Hall C, King S. Steele OG, et al. Eur J Neurosci. 2022 Nov;56(9):5476-5515. doi: 10.1111/ejn.15685. Epub 2022 Jun 6. Eur J Neurosci. 2022. PMID: 35510513 Free PMC article. Review. - Age-dependent effect of apolipoprotein E4 on functional outcome after controlled cortical impact in mice.
Mannix RC, Zhang J, Park J, Zhang X, Bilal K, Walker K, Tanzi RE, Tesco G, Whalen MJ. Mannix RC, et al. J Cereb Blood Flow Metab. 2011 Jan;31(1):351-61. doi: 10.1038/jcbfm.2010.99. Epub 2010 Jun 30. J Cereb Blood Flow Metab. 2011. PMID: 20588316 Free PMC article. - ApoE expression in macrophages communicates immunometabolic signaling that controls hyperlipidemia-driven hematopoiesis & inflammation via extracellular vesicles.
Phu TA, Ng M, Vu NK, Gao AS, Raffai RL. Phu TA, et al. J Extracell Vesicles. 2023 Aug;12(8):e12345. doi: 10.1002/jev2.12345. J Extracell Vesicles. 2023. PMID: 37593979 Free PMC article.
References
- Mahley R W. Science. 1988;240:622–630. - PubMed
- Reyland M E, Williams D L. J Biol Chem. 1991;266:21099–21104. - PubMed
- Ishigami M, Swertfeger D K, Granholm N A, Hui D Y. J Biol Chem. 1998;273:20156–20161. - PubMed
- Avila E M, Holdsworth G, Sasaki N, Jackson R L, Harmony J A K. J Biol Chem. 1982;257:5900–5909. - PubMed
- Hui D Y, Harmony J A K, Innerarity T L, Mahley R W. J Biol Chem. 1980;255:11775–11781. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous