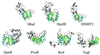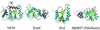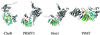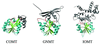AdoMet-dependent methylation, DNA methyltransferases and base flipping - PubMed (original) (raw)
Review
AdoMet-dependent methylation, DNA methyltransferases and base flipping
X Cheng et al. Nucleic Acids Res. 2001.
Abstract
Twenty AdoMet-dependent methyltransferases (MTases) have been characterized structurally by X-ray crystallography and NMR. These include seven DNA MTases, five RNA MTases, four protein MTases and four small molecule MTases acting on the carbon, oxygen or nitrogen atoms of their substrates. The MTases share a common core structure of a mixed seven-stranded beta-sheet (6 downward arrow 7 upward arrow 5 downward arrow 4 downward arrow 1 downward arrow 2 downward arrow 3 downward arrow) referred to as an 'AdoMet-dependent MTase fold', with the exception of a protein arginine MTase which contains a compact consensus fold lacking the antiparallel hairpin strands (6 downward arrow 7 upward arrow). The consensus fold is useful to identify hypothetical MTases during structural proteomics efforts on unannotated proteins. The same core structure works for very different classes of MTase including those that act on substrates differing in size from small molecules (catechol or glycine) to macromolecules (DNA, RNA and protein). DNA MTases use a 'base flipping' mechanism to deliver a specific base within a DNA molecule into a typically concave catalytic pocket. Base flipping involves rotation of backbone bonds in double-stranded DNA to expose an out-of-stack nucleotide, which can then be a substrate for an enzyme-catalyzed chemical reaction. The phenomenon is fully established for DNA MTases and for DNA base excision repair enzymes, and is likely to prove general for enzymes that require access to unpaired, mismatched or damaged nucleotides within base-paired regions in DNA and RNA. Several newly discovered MTase families in eukaryotes (DNA 5mC MTases and protein arginine and lysine MTases) offer new challenges in the MTase field.
Figures
Figure 1
Examples of DNA MTases (see Table 1). M._Hha_I (5mC), M._Hae_III (5mC), human DNMT2, M._Dpn_II (N6mA, α), M._Pvu_II (N4mC, β), M._Rsr_I (N6mA, β) and M._Taq_I (N6mA, γ). The AdoMet-dependent MTase fold is colored in green (β strands), cyan (α helices) and red (the loops after the carboxyl ends of β strands). The region(s) outside the MTase fold is colored in gray.
Figure 2
Examples of RNA MTases. VP39 (mRNA nucleoside-2′-O), ErmC′ or closely related ErmAM (rRNA N6mA), FtsJ and fibrillarin homolog.
Figure 3
Examples of protein MTases. CheR (glutamate-O), PRMT3 (arginine-N), Hmt1 (arginine-N) and PIMT (isoaspartate-O).
Figure 4
Examples of small molecule MTases: COMT (catechol-O), GNMT (glycine-N), and IOMT (isoflavone-O) or closely related ChOMT (chalcone-O).
Figure 5
Examples of DNA base flipping proteins (see Table 3) in complex with oligonucleotide containing an abasic site. (A) M._Hha_I (a DNA 5mC MTase), (B) human UDG, (C) human AAG, (D) E.coli endonuclease IV, (E) E.coli AlkA and (F) human HAP1. The protein is colored gray, the DNA is represented as a magenta stick model with the flipped abasic site in green, usually buried in a surface pocket in the protein.
Figure 6
30S ribosomal subunit flipped-out A1492 and A1493 from helix 44 of 16S RNA by binding of (left) paromomycin (seen in the difference electron density) and (right) initiation factor IF1 (in purple). The protein S12 is in orange, helix H44 in cyan.
Similar articles
- The conserved aspartate in motif III of b family AdoMet-dependent DNA methyltransferase is important for methylation.
Gopinath A, Kulkarni M, Ahmed I, Chouhan OP, Saikrishnan K. Gopinath A, et al. J Biosci. 2020;45:10. J Biosci. 2020. PMID: 31965988 - Crystal structure of the DpnM DNA adenine methyltransferase from the DpnII restriction system of streptococcus pneumoniae bound to S-adenosylmethionine.
Tran PH, Korszun ZR, Cerritelli S, Springhorn SS, Lacks SA. Tran PH, et al. Structure. 1998 Dec 15;6(12):1563-75. doi: 10.1016/s0969-2126(98)00154-3. Structure. 1998. PMID: 9862809 - Many paths to methyltransfer: a chronicle of convergence.
Schubert HL, Blumenthal RM, Cheng X. Schubert HL, et al. Trends Biochem Sci. 2003 Jun;28(6):329-35. doi: 10.1016/S0968-0004(03)00090-2. Trends Biochem Sci. 2003. PMID: 12826405 Free PMC article. Review. - Chemical Expansion of the Methyltransferase Reaction: Tools for DNA Labeling and Epigenome Analysis.
Vilkaitis G, Masevičius V, Kriukienė E, Klimašauskas S. Vilkaitis G, et al. Acc Chem Res. 2023 Nov 21;56(22):3188-3197. doi: 10.1021/acs.accounts.3c00471. Epub 2023 Oct 30. Acc Chem Res. 2023. PMID: 37904501 Free PMC article. - Closing in on human methylation-the versatile family of seven-β-strand (METTL) methyltransferases.
Falnes PØ. Falnes PØ. Nucleic Acids Res. 2024 Oct 28;52(19):11423-11441. doi: 10.1093/nar/gkae816. Nucleic Acids Res. 2024. PMID: 39351878 Free PMC article. Review.
Cited by
- Cell cycle regulated DNA methyltransferase: fluorescent tracking of a DNA strand-separation mechanism and identification of the responsible protein motif.
Konttinen O, Carmody J, Pathuri S, Anderson K, Zhou X, Reich N. Konttinen O, et al. Nucleic Acids Res. 2020 Nov 18;48(20):11589-11601. doi: 10.1093/nar/gkaa844. Nucleic Acids Res. 2020. PMID: 33053173 Free PMC article. - Structure analysis of Entamoeba histolytica DNMT2 (EhMeth).
Schulz EC, Roth HM, Ankri S, Ficner R. Schulz EC, et al. PLoS One. 2012;7(6):e38728. doi: 10.1371/journal.pone.0038728. Epub 2012 Jun 21. PLoS One. 2012. PMID: 22737219 Free PMC article. - On the evolutionary origin of eukaryotic DNA methyltransferases and Dnmt2.
Jurkowski TP, Jeltsch A. Jurkowski TP, et al. PLoS One. 2011;6(11):e28104. doi: 10.1371/journal.pone.0028104. Epub 2011 Nov 30. PLoS One. 2011. PMID: 22140515 Free PMC article. - Organization of the BcgI restriction-modification protein for the cleavage of eight phosphodiester bonds in DNA.
Smith RM, Marshall JJ, Jacklin AJ, Retter SE, Halford SE, Sobott F. Smith RM, et al. Nucleic Acids Res. 2013 Jan 7;41(1):391-404. doi: 10.1093/nar/gks1023. Epub 2012 Nov 11. Nucleic Acids Res. 2013. PMID: 23147005 Free PMC article. - The multistructural forms of box C/D ribonucleoprotein particles.
Yu G, Zhao Y, Li H. Yu G, et al. RNA. 2018 Dec;24(12):1625-1633. doi: 10.1261/rna.068312.118. Epub 2018 Sep 25. RNA. 2018. PMID: 30254138 Free PMC article. Review.
References
- Walsh C. (1979) Enzymatic Reaction Mechanisms. W.H. Freeman, San Francisco, CA, pp. 851–859.
- Dryden D.T.F. (1999) Bacterial DNA methyltransferases. In Cheng,X. and Blumenthal,R.M. (eds), S-Adenosylmethionine-Dependent Methyltransferases: Structures and Functions. World Scientific Publishing, NJ, pp. 283–340.
- Bestor T.H. (1988) Cloning of a mammalian DNA methyltransferase. Gene, 74, 9–12. - PubMed
- Yoder J.A. and Bestor,T.H. (1998) A candidate mammalian DNA methyltransferase related to pmt1p of fission yeast. Hum. Mol. Genet., 7, 279–284. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM61355/GM/NIGMS NIH HHS/United States
- GM46127/GM/NIGMS NIH HHS/United States
- GM49245/GM/NIGMS NIH HHS/United States
- R01 GM049245/GM/NIGMS NIH HHS/United States
- R01 GM061355/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases