A protein-protein interaction map of the Caenorhabditis elegans 26S proteasome - PubMed (original) (raw)
doi: 10.1093/embo-reports/kve184.
P Bello, N Thierry-Mieg, P Vaglio, J Hitti, L Doucette-Stamm, D Thierry-Mieg, J Reboul, S Boulton, A J Walhout, O Coux, M Vidal
Affiliations
- PMID: 11559592
- PMCID: PMC1084039
- DOI: 10.1093/embo-reports/kve184
A protein-protein interaction map of the Caenorhabditis elegans 26S proteasome
A Davy et al. EMBO Rep. 2001 Sep.
Abstract
The ubiquitin-proteasome proteolytic pathway is pivotal in most biological processes. Despite a great level of information available for the eukaryotic 26S proteasome-the protease responsible for the degradation of ubiquitylated proteins-several structural and functional questions remain unanswered. To gain more insight into the assembly and function of the metazoan 26S proteasome, a two-hybrid-based protein interaction map was generated using 30 Caenorhabditis elegans proteasome subunits. The results recapitulate interactions reported for other organisms and reveal new potential interactions both within the 19S regulatory complex and between the 19S and 20S subcomplexes. Moreover, novel potential proteasome interactors were identified, including an E3 ubiquitin ligase, transcription factors, chaperone proteins and other proteins not yet functionally annotated. By providing a wealth of novel biological hypotheses, this interaction map constitutes a framework for further analysis of the ubiquitin-proteasome pathway in a multicellular organism amenable to both classical genetics and functional genomics.
Figures
Fig. 1. PCR amplification of pORFs. PCR-amplified ORFs corresponding to potential C. elegans pORFs were analyzed by electrophoresis after organizing them by size in order to facilitate the read-out of the results (Reboul et al., 2001). PCRs were considered successful when a single band of the expected size was observed and accordingly, 30 out of 32 pORFs were successfully PCR amplified and cloned. The PCR product of Y110A7A.14 migrated as a longer fragment than its GeneFinder prediction, however this ORF was confirmed by sequencing (see Reboul et al., 2001 for an explanation). Two pORFs (F10G7.8 and F57B9.10) could not be Gateway cloned due to unsuccessful PCRs.
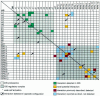
Fig. 2. Caenorhabditis elegans proteasome interaction mapping: two-hybrid interactions between 26S proteasome subunits. The data shown here was obtained for all pair-wise combinations by compiling both the matrix (DB-pORF/AD-pORF) and the screens (DB-pORF/AD-cDNAs). See Supplementary data for a detailed description of the comprehensive two-hybrid screens.
Fig. 3. Proteasome subunit/subunit interactions and new potential interactors. Circles and arrows represent proteins and two-hybrid interactions (from DB-X to AD-Y), respectively. (A) Comprehensive two-hybrid protein network of the C. elegans 26S proteasome. Red circles represent proteasome subunits and green circles represent other potential interactors. (B) Two-hybrid interactions between α and β subunits of the 20S. The α and β rings were separated and each subunit colored differently for clarity. (C) A new model for the 26S proteasome. The 19S subunits used in this study were colored in gray and interactions detected between 26S proteasome subunits are shown with red arrows. Novel potential interactors forming clusters with proteasome subunits and proteins previously characterized for which a model of interaction with the proteasome could be envisaged are shown in green.
Similar articles
- The 26S proteasome: ubiquitin-mediated proteolysis in the tunnel.
Kierszenbaum AL. Kierszenbaum AL. Mol Reprod Dev. 2000 Oct;57(2):109-10. doi: 10.1002/1098-2795(200010)57:2<109::AID-MRD1>3.0.CO;2-9. Mol Reprod Dev. 2000. PMID: 10984410 Review. - 20S proteasome activation promotes life span extension and resistance to proteotoxicity in Caenorhabditis elegans.
Chondrogianni N, Georgila K, Kourtis N, Tavernarakis N, Gonos ES. Chondrogianni N, et al. FASEB J. 2015 Feb;29(2):611-22. doi: 10.1096/fj.14-252189. Epub 2014 Nov 13. FASEB J. 2015. PMID: 25395451 Free PMC article. - Regulatory subunit interactions of the 26S proteasome, a complex problem.
Ferrell K, Wilkinson CR, Dubiel W, Gordon C. Ferrell K, et al. Trends Biochem Sci. 2000 Feb;25(2):83-8. doi: 10.1016/s0968-0004(99)01529-7. Trends Biochem Sci. 2000. PMID: 10664589 Review. - Reverse genetic analysis of the Caenorhabditis elegans 26S proteasome subunits by RNA interference.
Takahashi M, Iwasaki H, Inoue H, Takahashi K. Takahashi M, et al. Biol Chem. 2002 Jul-Aug;383(7-8):1263-6. doi: 10.1515/BC.2002.140. Biol Chem. 2002. PMID: 12437114 - Identification of ubiquitin-like protein-binding subunits of the 26S proteasome.
Saeki Y, Sone T, Toh-e A, Yokosawa H. Saeki Y, et al. Biochem Biophys Res Commun. 2002 Aug 30;296(4):813-9. doi: 10.1016/s0006-291x(02)02002-8. Biochem Biophys Res Commun. 2002. PMID: 12200120
Cited by
- A genomewide screen for suppressors of par-2 uncovers potential regulators of PAR protein-dependent cell polarity in Caenorhabditis elegans.
Labbé JC, Pacquelet A, Marty T, Gotta M. Labbé JC, et al. Genetics. 2006 Sep;174(1):285-95. doi: 10.1534/genetics.106.060517. Epub 2006 Jul 2. Genetics. 2006. PMID: 16816419 Free PMC article. - Development of Small Molecular Proteasome Inhibitors Using a Caenorhabditis elegans Screen.
Nayak S, Fiaschi M, King D, Tabakin ER, Wood L, Hunt DA. Nayak S, et al. Int J Med Chem. 2014;2014:237286. doi: 10.1155/2014/237286. Epub 2014 Nov 11. Int J Med Chem. 2014. PMID: 25436151 Free PMC article. - A new pooling strategy for high-throughput screening: the Shifted Transversal Design.
Thierry-Mieg N. Thierry-Mieg N. BMC Bioinformatics. 2006 Jan 19;7:28. doi: 10.1186/1471-2105-7-28. BMC Bioinformatics. 2006. PMID: 16423300 Free PMC article. - The ubiquitin proteasome system in Caenorhabditis elegans and its regulation.
Papaevgeniou N, Chondrogianni N. Papaevgeniou N, et al. Redox Biol. 2014 Jan 18;2:333-47. doi: 10.1016/j.redox.2014.01.007. eCollection 2014. Redox Biol. 2014. PMID: 24563851 Free PMC article. Review. - Specific SKN-1/Nrf stress responses to perturbations in translation elongation and proteasome activity.
Li X, Matilainen O, Jin C, Glover-Cutter KM, Holmberg CI, Blackwell TK. Li X, et al. PLoS Genet. 2011 Jun;7(6):e1002119. doi: 10.1371/journal.pgen.1002119. Epub 2011 Jun 9. PLoS Genet. 2011. PMID: 21695230 Free PMC article.
References
- Bays N.W., Gardner, R.G., Seelig, L.P., Joazeiro, C.A. and Hampton, R.Y. (2001) Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nature Cell Biol., 3, 24–29. - PubMed
- Cagney G., Uetz, P. and Fields, S. (2001) Two-hybrid analysis of the Saccharomyces cerevisiae 26S proteasome. Physiolog. Genomics, in press. - PubMed
- Ciechanover A., Orian, A. and Schwartz, A.L. (2000) Ubiquitin-mediated proteolysis: biological regulation via destruction. BioEssays, 97, 2497–2502. - PubMed
- Durbin R. and Thierry-Mieg, J. (1994) The ACeDB genome database. In Suhai, S. (ed.), Computational Methods in Genome Research. Plenum Press, New York, NY.
Publication types
MeSH terms
Substances
Grants and funding
- P01 CA080111/CA/NCI NIH HHS/United States
- R01 HG001715/HG/NHGRI NIH HHS/United States
- 232/PHS HHS/United States
- 5R01HG01715-02/HG/NHGRI NIH HHS/United States
- 7R33 CA81658-02/CA/NCI NIH HHS/United States
- P01CA80111-02/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases