The xeroderma pigmentosum group E gene product DDB2 is a specific target of cullin 4A in mammalian cells - PubMed (original) (raw)
The xeroderma pigmentosum group E gene product DDB2 is a specific target of cullin 4A in mammalian cells
A Nag et al. Mol Cell Biol. 2001 Oct.
Abstract
The damaged-DNA binding protein DDB consists of two subunits, DDB1 (127 kDa) and DDB2 (48 kDa). Mutations in the DDB2 subunit have been detected in patients suffering from the repair deficiency disease xeroderma pigmentosum (group E). In addition, recent studies suggested a role for DDB2 in global genomic repair. DDB2 also exhibits transcriptional activity. We showed that expression of DDB1 and DDB2 stimulated the activity of the cell cycle regulatory transcription factor E2F1. Here we show that DDB2 is a cell cycle-regulated protein. It is present at a low level in growth-arrested primary fibroblasts, and after release the level peaks at the G(1)/S boundary. The cell cycle regulation of DDB2 involves posttranscriptional mechanisms. Moreover, we find that an inhibitor of 26S proteasome increases the level of DDB2, suggesting that it is regulated by the ubiquitin-proteasome pathway. Our previous study indicated that the cullin family protein Cul-4A associates with the DDB2 subunit. Because cullins are involved in the ubiquitin-proteasome pathway, we investigated the role of Cul-4A in regulating DDB2. Here we show that DDB2 is a specific target of Cul-4A. Coexpression of Cul-4A, but not Cul-1 or other highly related cullins, increases the ubiquitination and the decay rate of DDB2. A naturally occurring mutant of DDB2 (2RO), which does not bind Cul-4A, is not affected by coexpression of Cul-4A. Studies presented here identify a specific function of the Cul-4A gene, which is amplified and overexpressed in breast cancers.
Figures
FIG. 1
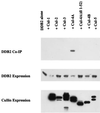
DDB2 specifically binds Cul-4A. Plasmids expressing V5-tagged cullins (19 μg of cullin 1, 2, 3, or 5; 3 μg of the wild-type or mutant cullin 4A; or 6 μg of cullin 4B DNA) were transfected in human 293 cells along with a plasmid expressing T7-tagged DDB2 (1 μg). Total DNA was made up to 20 μg with empty V5 vector where necessary. The transfected cells were washed with PBS, and cell extracts were prepared as described in Materials and Methods. The extracts were subjected to immunoprecipitation with V5 antibody. The immunoprecipitates were eluted with gel-loading buffer at room temperature for 10 min, boiled, and subjected to a Western blot assay. The blot was probed with T7 antibody to assay for the presence of coimmuoprecipitated DDB2.
FIG. 2
DDB2 is regulated during the cell cycle by a posttranscriptional mechanism. (A) Human primary fibroblasts were synchronized by serum starvation for 72 h followed by stimulation with 10% FBS. At the indicated time points, cells were harvested and nuclear extracts were prepared and subjected to Western blot analysis (with 250 μg of nuclear extract) for DDB2, Cul-4A, cyclin E, and cdk2. The Lower panel shows the changes in the levels of DDB2 and cyclophilin mRNA at various time points of the cell cycle. Total RNA from the cells was subjected to RNase protection assays using antisense RNA probes corresponding to DDB2 and cyclophilin as described in Materials and Methods. (B) Plot showing the relative levels of the DDB2 protein and mRNA during the progression of the cell cycle. Band intensities were determined by densitometric scanning of the films corresponding to the Western blot and RNase protection assays. After normalization, the relative intensities were plotted.
FIG. 3
DDB2 accumulates in the nucleus at the G1/S boundary in synchronized population of HeLa cells, and its level is regulated by the ubiquitin-proteasome pathway. HeLa cells were arrested at the G1/S boundary by a thymidine-aphidicolin double block. The cells were harvested at indicated time points, and nuclear extracts were prepared as described in Materials and Methods and analyzed for the levels of DDB2 (100 μg of extract), cyclin A (50 μg), and cdk2 (50 μg) by a Western blot assay. The lower panel shows the effect of proteasome inhibitor on the level of DDB2. For this, HeLa suspension culture cells were treated with either MG 132 (10 μM) or equal volume of dimethyl sulfoxide for 5 h. Cells were harvested, and 100 μg of nuclear extracts was analyzed for DDB2.
FIG. 4
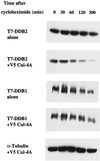
Coexpression of Cul-4A specifically reduces the half-life of DDB2. Five plates (10-cm-diameter dish) of HeLa cells were transfected with plasmids that express T7 epitope-tagged DDB1 and DDB2 in the presence or absence of a plasmid expressing V5-tagged Cul-4A. At 16 h posttransfection, the cells were trypsinized and the cells from the same set were pooled and replated on five plates. After 24 h, cycloheximide was added to the medium at a final concentration of 20 μg/ml and the cells were harvested at the indicated time points. Total-cell extracts were prepared as described in Materials and Methods. Then 100-μg portions of the transfected cell extracts were analyzed by Western blot assays. The blot was probed with antibodies against the T7 epitope.
FIG. 5
(A) The decay rate of DDB2 is specifically enhanced by the expression of Cul-4A. Five plates of HeLa cells were transfected with plasmids that express T7 epitope-tagged DDB2 alone or with plasmids expressing V5-tagged Cul-4A or V5-Cul-1 or V5-Cul-4A (dl 1–52) or Cul-4B. At 16 h posttransfection, the cells from same set were trypsinized, pooled, and replated on five plates. After 24 h, the cells were treated with cycloheximide (20 μg/ml) and harvested at the indicated time points. Then 100-μg portions of the transfected cell extracts were analyzed by Western blot assays. The blot was probed with antibodies against the T7 epitope. (B) Plot showing the decay rate of DDB2 in the presence of different cullins. The band intensities were determined by densitometric scanning. For each plot, the band intensity corresponding to the zero hour time point was taken as 100% and the relative intensities of the bands at various time points were plotted. Symbols: ⧫, DDB2; ■, DDB2 + Cul-1; ▵, DDB2 + Cul4A; ×, DDB2 + Cul-4B; ∗, DDB2 + Cul-4A (dl 1–52).
FIG. 6
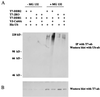
(A) Cul-4A induces ubiqitination of DDB2. HeLa cells were transfected with plasmids expressing T7-DDB2 (4 μg), T7-DDB1 (6 μg), T7-2RO (4 μg), His-tagged ubiquitin (U6) (1 μg), and V5-Cul-4A (9 μg) in the indicated combinations. At 40 h after transfection, the cells were treated with MG 132 for 5 h. The cells were washed and lysed as described in Materials and Methods. Equal amount of extracts (1 mg) were immunoprecipitated (IP) with T7 antibodies (ab) covalently linked to agarose beads, and the immunoprecipitates were subjected to Western blot assay. The blot was probed with monoclonal antibodies against ubiquitin. (B) Western blot of the extracts (100 μg) probed with T7 antibody (ab) to assay for the expression levels of DDB2 in the above extracts.
FIG. 7
DDB2 itself is ubiquitinated by Cul-4A. HeLa cells were transfected with plasmids expressing T7-tagged DDB2 and V5-tagged DDB1 in the presence and absence of the Cul-4A expression plasmid. At 40 h after transfection, the cells were treated with MG 132 for 5 h. Total-cell extracts were prepared as described in Materials and Methods. Equal amount of the transfected cell extracts were treated with 2% SDS (final concentration) at 37°C for 10 min to disrupt protein-protein interactions. The SDS-treated extracts were diluted 10-fold and then subjected to immunoprecipitation (IP) with T7 antibody covalently linked to agarose beads. The immunoprecipitates were subjected to Western blot assay by using monoclonal antibodies against T7 for DDB2 and V5 for DDB1 (A) and ubiquitin (Ub ab) for ubiquitinated proteins (B).
FIG. 8
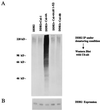
DDB2 is a specific ubiquitination target of Cul-4A. (A) Cells were transfected with plasmids expressing T7-DDB2 or His-tagged ubiquitin in combination with empty vector, V5-Cul-1, V5-Cul-4A, V5-Cul-4A(dl 1–52), or V5-Cul-4B. At 40 h after transfection, the cells were treated with MG 132 for 5 h. Total-cell extracts were prepared as described in Materials and Methods. Equal amount of extracts were treated with 2% SDS at 37°C for 10 min. The SDS-treated extracts were diluted 10-fold and then subjected to immunoprecipitation (IP) with T7 antibodies and Western blot assay with monoclonal antibodies against ubiquitin (Ub-ab). (B) Level of expression of T7-DDB2 in the extracts. Transfected cell extracts (100 μg) were probed with T7- antibody in a Western blot to assay for DDB2 expression.
Similar articles
- Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.
Salisbury L, Baraitser L. Salisbury L, et al. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review. - "I've Spent My Whole Life Striving to Be Normal": Internalized Stigma and Perceived Impact of Diagnosis in Autistic Adults.
Huang Y, Trollor JN, Foley KR, Arnold SRC. Huang Y, et al. Autism Adulthood. 2023 Dec 1;5(4):423-436. doi: 10.1089/aut.2022.0066. Epub 2023 Dec 12. Autism Adulthood. 2023. PMID: 38116050 Free PMC article. - Identification of a novel toxicophore in anti-cancer chemotherapeutics that targets mitochondrial respiratory complex I.
Stephenson ZA, Harvey RF, Pryde KR, Mistry S, Hardy RE, Serreli R, Chung I, Allen TE, Stoneley M, MacFarlane M, Fischer PM, Hirst J, Kellam B, Willis AE. Stephenson ZA, et al. Elife. 2020 May 20;9:e55845. doi: 10.7554/eLife.55845. Elife. 2020. PMID: 32432547 Free PMC article. - Using Experience Sampling Methodology to Capture Disclosure Opportunities for Autistic Adults.
Love AMA, Edwards C, Cai RY, Gibbs V. Love AMA, et al. Autism Adulthood. 2023 Dec 1;5(4):389-400. doi: 10.1089/aut.2022.0090. Epub 2023 Dec 12. Autism Adulthood. 2023. PMID: 38116059 Free PMC article. - Trends in Surgical and Nonsurgical Aesthetic Procedures: A 14-Year Analysis of the International Society of Aesthetic Plastic Surgery-ISAPS.
Triana L, Palacios Huatuco RM, Campilgio G, Liscano E. Triana L, et al. Aesthetic Plast Surg. 2024 Oct;48(20):4217-4227. doi: 10.1007/s00266-024-04260-2. Epub 2024 Aug 5. Aesthetic Plast Surg. 2024. PMID: 39103642 Review.
Cited by
- The emerging role for Cullin 4 family of E3 ligases in tumorigenesis.
Cheng J, Guo J, North BJ, Tao K, Zhou P, Wei W. Cheng J, et al. Biochim Biophys Acta Rev Cancer. 2019 Jan;1871(1):138-159. doi: 10.1016/j.bbcan.2018.11.007. Epub 2018 Dec 30. Biochim Biophys Acta Rev Cancer. 2019. PMID: 30602127 Free PMC article. Review. - The deubiquitinating protein USP24 interacts with DDB2 and regulates DDB2 stability.
Zhang L, Lubin A, Chen H, Sun Z, Gong F. Zhang L, et al. Cell Cycle. 2012 Dec 1;11(23):4378-84. doi: 10.4161/cc.22688. Epub 2012 Nov 16. Cell Cycle. 2012. PMID: 23159851 Free PMC article. - Transcriptional and Posttranslational Regulation of Nucleotide Excision Repair: The Guardian of the Genome against Ultraviolet Radiation.
Park JM, Kang TH. Park JM, et al. Int J Mol Sci. 2016 Nov 4;17(11):1840. doi: 10.3390/ijms17111840. Int J Mol Sci. 2016. PMID: 27827925 Free PMC article. Review. - CRL1-FBXO11 promotes Cdt2 ubiquitylation and degradation and regulates Pr-Set7/Set8-mediated cellular migration.
Abbas T, Mueller AC, Shibata E, Keaton M, Rossi M, Dutta A. Abbas T, et al. Mol Cell. 2013 Mar 28;49(6):1147-58. doi: 10.1016/j.molcel.2013.02.003. Epub 2013 Mar 7. Mol Cell. 2013. PMID: 23478445 Free PMC article. - Nucleotide Excision Repair: Finely Tuned Molecular Orchestra of Early Pre-incision Events.
Zhu Q, Wani AA. Zhu Q, et al. Photochem Photobiol. 2017 Jan;93(1):166-177. doi: 10.1111/php.12647. Epub 2016 Nov 17. Photochem Photobiol. 2017. PMID: 27696486 Free PMC article. Review.
References
- Abramic M, Levine A S, Protic M. Purification of an ultraviolet-inducible, damage-specific DNA-binding protein from primate cells. J Biol Chem. 1991;266:22439–22500. - PubMed
- Bai C, Sen P, Mathias N, Hofmann K, Goebl M, Harper J W, Elledge S J. SKP1 connects cell cycle regulation to ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. - PubMed
- Chen L-C, Manjeshwar S, Lu Y, Moore D, Ljung B-M, Kuo W-L, Dairkee S H, Wernick M, Colins C, Smith H S. The human homologue for the Caenorhabditis elegans Cul-4 gene is amplified and overexpressed in primary breast cancers. Cancer Res. 1998;58:3677–3683. - PubMed
- Cho R J, Huang M, Campbell M J, Dong H, Steinmetz L, Sapinoso L, Hampton G, Elledge S J, Davis R W, Lockhart D J. Transcriptional regulation and function during the human cell cycle. Nat Genet. 2001;27:48–54. - PubMed
- Chu G, Chang E. Xeroderma pigmentosum group E cells lack a nuclear factor that binds to damaged DNA. Science. 1988;242:564–567. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources