Widespread use of TATA elements in the core promoters for RNA polymerases III, II, and I in fission yeast - PubMed (original) (raw)
Widespread use of TATA elements in the core promoters for RNA polymerases III, II, and I in fission yeast
M Hamada et al. Mol Cell Biol. 2001 Oct.
Abstract
In addition to directing transcription initiation, core promoters integrate input from distal regulatory elements. Except for rare exceptions, it has been generally found that eukaryotic tRNA and rRNA genes do not contain TATA promoter elements and instead use protein-protein interactions to bring the TATA-binding protein (TBP), to the core promoter. Genomewide analysis revealed TATA elements in the core promoters of tRNA and 5S rRNA (Pol III), U1 to U5 snRNA (Pol II), and 37S rRNA (Pol I) genes in Schizosaccharomyces pombe. Using tRNA-dependent suppression and other in vivo assays, as well as in vitro transcription, we demonstrated an obligatory requirement for upstream TATA elements for tRNA and 5S rRNA expression in S. pombe. The Pol III initiation factor Brf is found in complexes with TFIIIC and Pol III in S. pombe, while TBP is not, consistent with independent recruitment of TBP by TATA. Template commitment assays are consistent with this and confirm that the mechanisms of transcription complex assembly and initiation by Pol III in S. pombe differ substantially from those in other model organisms. The results were extended to large-rRNA synthesis, as mutation of the TATA element in the Pol I promoter also abolishes rRNA expression in fission yeast. A survey of other organisms' genomes reveals that a substantial number of eukaryotes may use widespread TATAs for transcription. These results indicate the presence of TATA-unified transcription systems in contemporary eukaryotes and provide insight into the residual need for TBP by all three Pols in other eukaryotes despite a lack of TATA elements in their promoters.
Figures
FIG. 1
Genomewide analysis reveals TATA motifs upstream of S. pombe tRNA genes. (A) Sequence Logos of the genomic 5′ flanking regions of tRNA sequences at 174 S. pombe, 275 S. cerevisiae, 425 D. melanogaster, 625 H. sapiens, and 600 A. thaliana loci. The positions of the A and B box promoter elements are indicated above the Logos. Note the relatively low level of sequence information content in the S. pombe A box, at the no. 10 position indicated by the asterisk, compared to that of S. cerevisiae. Although the full sets of sequences were used for the upstream regions and the first half of the tRNA for all species, for H. sapiens, A. thaliana, and D. melanogaster, the B box and downstream regions were limited to 200, 215, and 250 sequences, respectively, for practical reasons. Approximately 20% of the H. sapiens sequences were identified by tRNAscan-SE as “possible pseudogenes,” probably reflective of tRNA-like repetitive elements in the human genome (42). Approximately 1% each of the A. thaliana, S. pombe, and D. melanogaster sequences and none of the S. cerevisiae sequences were identified as possible pseudogenes. Common features of tRNA structure are indicated below the Logos. M1 indicates the first base of mature tRNA. A gap representing introns and sequences extending through the variable stem-loop (not shown) is designated i/v. ac, anticodon. (B) Sequence Logo of the S. pombe and A. thaliana sequences after prealignment of the upstream regions by the Clustal program. The numbering under the upstream region is relative to the putative consensus +1 site.
FIG. 2
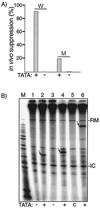
Upstream TATA is a positive determinant of tRNA expression in vivo and in a homologous in vitro S. pombe system. (A) The TATATA motifs of two opal suppressor tRNA genes, tRNAserUGA-W (W) and tRNAserUGA-M (M), were left unchanged (+) or replaced with GGATCC (−) as indicated along the horizontal axis and examined for suppressor activity in vivo as previously described (16, 24). (B) In vitro transcription in an _S. pombe_-derived extract was performed using three tRNA genes, containing (+) or lacking (−) upstream TATA elements, and empty plasmid control (c) as indicated below the lanes. Lanes 1 and 2, _S. cerevisiae_-derived tRNAserUCA gene; lanes 3 and 4, _S. pombe_-derived tRNAserUGA-M gene; lane 5, control plasmid containing no tRNA gene; lane 6, _S. pombe_-derived tRNAserUGA-M-3T gene (produces longer transcript [16]). The S. cerevisiae and S. pombe genes differ in size due to a 15-nt intron in the latter, and their A and B box elements are identical except at one position, G10 in the former and T10 in the latter. Transcript bands are indicated by arrows. -RM indicates a recovery marker added to the reactions, and -IC indicates an extract-derived internal control.
FIG. 3
Essential TATA motifs upstream of S. pombe 5S rRNA genes. (A) Sequence Logo of the upstream regions of multiple dispersed S. pombe 5S rDNA loci. The first base (G) of mature 5S rRNA was included as the last base on the right. (B) Northern blot analysis. A neutral sequence tag allows detection of the tagged transcript, designated 5S* rRNA, which is distinguishable from endogenous 5S rRNA. In vivo expression of plasmid-borne TATA-containing (+) and TATA-less (−) 5S* rRNA genes was monitored with a 5S*-specific probe. C, control. (C) The blot in panel B was stripped and rehybridized to detect endogenous 5S rRNA.
FIG. 4
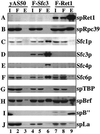
Fission yeast Brf and B" associate with Pol III TF complexes in the absence of stably associated TBP. Extracts prepared from a wild-type strain, the FLAG-Sfc3 strain, and a FLAG-spRet1p strain were incubated with anti-FLAG immunoglobulin G (M2)-agarose. After incubation, the supernatants were collected as the flowthrough, the agarose was washed five times with buffer containing 250 mM NaCl, and the bound material was eluted. The input (I), flowthrough (F), and eluate (E) of the M2-agarose from the three extracts were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and analyzed by immunoblotting using antisera to the proteins as indicated on the right.
FIG. 5
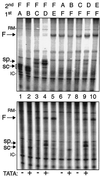
An upstream TATA is required to program a functional Pol III preinitiation complex on an A box- and B box-containing tRNA gene in S. pombe. Transcription complexes were monitored using template exclusion assays performed in parallel in S. cerevisiae (top) and S. pombe (bottom) in vitro transcription systems. The tRNA genes used for Fig. 2B were tested for the ability to form stable transcription complexes. After incubation of the first template (the order of addition is indicated above the lanes) with transcription buffer (previously determined to be optimal at 20 min), nucleoside triphosphates and [α-32P]GTP were added along with the second template (indicated above the lanes), and transcription was allowed to proceed. The reaction was stopped, and RNA was extracted and separated on 6% polyacrylamide–8 M urea. The templates were as follows: A, sc-tRNASer −TATA; B, sc-tRNASer +TATA; C, sp-tRNASer −TATA; D, sp-tRNASer +TATA; E, pJK148 (empty vector); F, sp-tRNASer-3T +TATA. The arrows point to the different transcripts synthesized.
FIG. 6
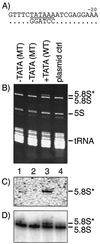
Pol I-transcribed large-rRNA genes use an upstream TATA motif in the core promoter region. (A) Sequence of promoter region (−40 through −20 relative to +1), with TATAAA motif underlined, of S. pombe rDNA (top line) (8) and TATA mutant (bottom line); dots indicate identical residues. (B) Ethidium bromide-stained polyacrylamide gel. Total RNA was isolated from S. pombe strains that had been transformed with plasmids containing 37S rRNA genes in which the 5.8S rRNA sequence contained a unique sequence tag (5.8S*; see the text). Lanes 1 and 2, two independent isolates in which the TATA element was mutated by site-directed substitution; lane 3, wild-type TATA; lane 4, control plasmid containing no rRNA gene. (C) Northern blot analysis using oligonucleotide probe conditions specific for the tagged 5.8* rRNA; the lanes are the same as in panel B. (D) The Northern blot in panel B was stripped and hybridized using an oligonucleotide probe that recognizes both wild-type (endogenous) 5.8S rRNA and 5.8* rRNA; the lanes are the same as in panel B.
FIG. 7
Conserved TATA motifs in the promoters of small RNA genes in S. pombe. (A) Pol III-transcribed genes. The upstream TATA, internal control region (ICR) (for 5S only), A box and B box (for tRNAs and U6 and 7SL RNAs), and terminators (T) are shown. The bent arrows indicate the +1 positions (as previously determined). Consensus sequences for the A and B box elements that fit all tRNA genes, as well as U6 and 7SL genes, are shown below the schematics; boldface letters are invariant or very highly conserved. The U6 B box resides in an intron (shaded; see the text) (B) Upstream regions of Pol II-transcribed snRNA genes are presented both in Logo (above) and as individual aligned sequences, ending at +1.
Similar articles
- The fission yeast TFIIB-related factor limits RNA polymerase III to a TATA-dependent pathway of TBP recruitment.
Huang Y, McGillicuddy E, Weindel M, Dong S, Maraia RJ. Huang Y, et al. Nucleic Acids Res. 2003 Apr 15;31(8):2108-16. doi: 10.1093/nar/gkg301. Nucleic Acids Res. 2003. PMID: 12682361 Free PMC article. - Comparison of the RNA polymerase III transcription machinery in Schizosaccharomyces pombe, Saccharomyces cerevisiae and human.
Huang Y, Maraia RJ. Huang Y, et al. Nucleic Acids Res. 2001 Jul 1;29(13):2675-90. doi: 10.1093/nar/29.13.2675. Nucleic Acids Res. 2001. PMID: 11433012 Free PMC article. Review. - Differential mode of TBP utilization in transcription of the tRNA[Ser]Sec gene and TATA-less class III genes.
Park JM, Lee JY, Hatfield DL, Lee BJ. Park JM, et al. Gene. 1997 Sep 1;196(1-2):99-103. doi: 10.1016/s0378-1119(97)00211-4. Gene. 1997. PMID: 9322746 - Regulation of snRNA gene expression by the Drosophila melanogaster small nuclear RNA activating protein complex (DmSNAPc).
Hung KH, Stumph WE. Hung KH, et al. Crit Rev Biochem Mol Biol. 2011 Feb;46(1):11-26. doi: 10.3109/10409238.2010.518136. Epub 2010 Oct 6. Crit Rev Biochem Mol Biol. 2011. PMID: 20925482 Review.
Cited by
- tRNAomics: tRNA gene copy number variation and codon use provide bioinformatic evidence of a new anticodon:codon wobble pair in a eukaryote.
Iben JR, Maraia RJ. Iben JR, et al. RNA. 2012 Jul;18(7):1358-72. doi: 10.1261/rna.032151.111. Epub 2012 May 14. RNA. 2012. PMID: 22586155 Free PMC article. - Genomic organization of human transcription initiation complexes.
Venters BJ, Pugh BF. Venters BJ, et al. Nature. 2013 Oct 3;502(7469):53-8. doi: 10.1038/nature12535. Epub 2013 Sep 18. Nature. 2013. PMID: 24048476 Free PMC article. Retracted. - TATA-Like Boxes in RNA Polymerase III Promoters: Requirements for Nucleotide Sequences.
Tatosyan KA, Stasenko DV, Koval AP, Gogolevskaya IK, Kramerov DA. Tatosyan KA, et al. Int J Mol Sci. 2020 May 25;21(10):3706. doi: 10.3390/ijms21103706. Int J Mol Sci. 2020. PMID: 32466110 Free PMC article. - Nucleotide Context Can Modulate Promoter Strength in Genes Transcribed by RNA Polymerase III.
Stasenko DV, Tatosyan KA, Borodulina OR, Kramerov DA. Stasenko DV, et al. Genes (Basel). 2023 Mar 27;14(4):802. doi: 10.3390/genes14040802. Genes (Basel). 2023. PMID: 37107560 Free PMC article. - Prp4 Kinase Grants the License to Splice: Control of Weak Splice Sites during Spliceosome Activation.
Eckert D, Andrée N, Razanau A, Zock-Emmenthal S, Lützelberger M, Plath S, Schmidt H, Guerra-Moreno A, Cozzuto L, Ayté J, Käufer NF. Eckert D, et al. PLoS Genet. 2016 Jan 5;12(1):e1005768. doi: 10.1371/journal.pgen.1005768. eCollection 2016 Jan. PLoS Genet. 2016. PMID: 26730850 Free PMC article.
References
- Bogenhagen D F, Wormington W M, Brown D D. Stable transcription complexes of Xenopus 5S RNA genes: a means to maintain the differentiated state. Cell. 1982;28:413–421. - PubMed
- Burke T W, Willy P J, Kutach A K, Butler J E, Kadonaga J T. The DPE, a conserved downstream core promoter element that is functionally analogous to the TATA box. Cold Spring Harbor Symp Quant Biol. 1998;63:75–82. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources