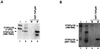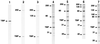Positive and negative TAF(II) functions that suggest a dynamic TFIID structure and elicit synergy with traps in activator-induced transcription - PubMed (original) (raw)
Positive and negative TAF(II) functions that suggest a dynamic TFIID structure and elicit synergy with traps in activator-induced transcription
M Guermah et al. Mol Cell Biol. 2001 Oct.
Abstract
Human transcription factor TFIID contains the TATA-binding protein (TBP) and several TBP-associated factors (TAF(II)s). To elucidate the structural organization and function of TFIID, we expressed and characterized the product of a cloned cDNA encoding human TAF(II)135 (hTAF(II)135). Comparative far Western blots have shown that hTAF(II)135 interacts strongly with hTAF(II)20, moderately with hTAF(II)150, and weakly with hTAF(II)43 and hTAF(II)250. Consistent with these observations and with sequence relationships of hTAF(II)20 and hTAF(II)135 to histones H2B and H2A, respectively, TFIID preparations that contain higher levels of hTAF(II)135 also contain higher levels of hTAF(II)20, and the interaction between hTAF(II)20 and hTAF(II)135 is critical for human TFIID assembly in vitro. From a functional standpoint, hTAF(II)135 has been found to interact strongly and directly with hTFIIA and (within a complex that also contains hTBP and hTAF(II)250) to specifically cooperate with TFIIA to relieve TAF(II)250-mediated repression of TBP binding and function on core promoters. Finally, we report a functional synergism between TAF(II)s and the TRAP/Mediator complex in activated transcription, manifested as hTAF(II)-mediated inhibition of basal transcription and a consequent TRAP requirement for both a high absolute level of activated transcription and a high and more physiological activated/basal transcription ratio. These results suggest a dynamic TFIID structure in which the switch from a basal hTAF(II)-enhanced repression state to an activator-mediated activated state on a promoter may be mediated in part through activator or coactivator interactions with hTAF(II)135.
Figures
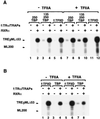
FIG. 1
Comparative far Western assays showing strong interactions between TAFII135 and hTAFII20. (A) Individually expressed (Sf9 cells via baculovirus vectors) and purified TFIID subunits were resolved on SDS-PAGE and visualized by Coomassie staining, M, protein markers, with molecular sizes indicated in kilodaltons on the left. (B) A set of gels equivalent to those in panel A were transferred onto nitrocellulose, denatured-renatured, and used for binding with 32P-labeled and purified hTAFII135. After extensive washing, bound material was analyzed by autoradiography.
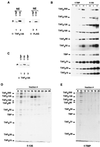
FIG. 2
The extreme C-terminal portion of hTAFII135 interacts with hTAFII20. The indicated deletion mutants of hTAFII135 were [35S]methionine-labeled and incubated with GST-hTAFII20. Input samples (I) contained 10% of the amount used for binding (B). The arrow indicates the band corresponding to the appropriate mutant hTAFII135 deletion.
FIG. 3
At least two molecules of TAFII135 coexist in TFIID. (A) Western blot analysis was performed using anti-hTAFII135 (lanes 1 and 2) and anti-Flag antibodies (lanes 3 and 4) to probe SDS-PAGE-resolved proteins in nuclear extracts. Both the endogenous hTAFII135 (open arrow) and Flag-TAFII135 (solid) are present in nuclear extract (NE) prepared from the Flag-TAFII135 (f:135) cell line (lane 1), whereas only the endogenous hTAFII135 band is present (lane 2) in NE from the Flag-TBP (f:TBP) cell line. (B) Comparison of TBP and TAFII expression in the TFIID purified from f:TBP and f:135 cell lines. As indicated, increasing amounts (microliters) of purified TFIID from either cell line were analyzed by Western blotting with antibodies generated against each indicated TFIID subunit. (C) Western blot analysis was performed using anti-hTAFII135 antibody. The endogenous hTAFII135 (open arrow) and Flag-hTAFII135 (solid arrow) proteins coexist in the purified TFIID prepared from the f:135 cell line (lane 2). In the TFIID purified from the f:TBP cell line, only the endogenous hTAFII135 (open arrow) is detected (lanes 1 and 3). (D and E) Indicated fractions, obtained from gel filtration on Superose 6, of purified TFIID from f:TBP and f:135 cell lines, respectively, were resolved by SDS-PAGE and silver stained.
FIG. 4
Role of TAFII135-hTAFII20 interactions in human TFIID assembly. Partially reconstituted TFIID species were resolved by SDS-PAGE and visualized by silver staining. The complexes were assembled with purified subunits that were individually expressed in Sf9 cells via baculovirus vectors (see text). Lane 1 contained TBP alone. Complexes analyzed in other lanes contained hTBP-hTAFII250 (lane 2), hTBP-TAFII250-TAFII135 (lane 3), hTBP-TAFII250-TAFII80-TAFII31, to which added TAFII135 failed to bind in a stoichiometric manner (lane 4), hTBP-TAFII250-TAFII80-TAFII31-hTAFII20 (lane 5), hTBP-TAFII250-hTAFII135-hTAFII80-hTAFII31-hTAFII20 (lane 6), and hTBP-TAFII250-hTAFII135-hTAFII100-hTAFII80-hTAFII31-hTAFII20 (lane 7).
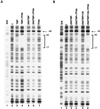
FIG. 5
Strong in vitro interactions between hTAFII135 and TFIIA subunits. (A) Equivalent amounts of full-length [35S]methionine-labeled hTAFII135 were incubated with the indicated GST derivatives (1 μg of each). After extensive washing, bound proteins were resolved by SDS-PAGE and analyzed by autoradiography. Input samples contained 10% of the amount used for binding. The arrow indicates the band corresponding to full length hTAFII135. (B) Deletion mutants of hTAFII135 that were generated, [35S]methionine labeled, and subsequently used for interaction studies (C and D). Equivalent amounts of [35S]methionine-labeled full length hTAFII135 and different deletion mutants were incubated with either GST-IIAp55 subunit (C) or GST-IIAp12 subunit (D). After washing, bound material was processed as in A.
FIG. 6
hTAFII135 and TFIIA synergize to specifically relieve the hTAFII250-mediated inhibition of TBP binding. DNase I footprinting of hTBP, hTBP-hTAFII250, and hTBP-TAFII250-TAFII135. The transcribed DNA template spanning positions −91 to +85 of the adenovirus major late promoter was prepared. The Maxam-Gilbert sequencing method was used to prepare the G/A footprinting marker (lanes A1 and B1). No protein (lanes A2 and B2), hTBP (2.5 ng for lanes A2 and A3), hTBP-hTAFII250 (10 ng of TBP content for lanes A5, A6, B3, and B4), hTBP-TAFII250-TAFII135 (10 ng of TBP content for lanes B5 and B6). Purified TFIIA was added to reactions corresponding to lanes A4, A6, A7, B4, and B6.
FIG. 7
Effect of TFIIA on basal and TRα-TRAP-activated transcription in the presence of TFIID and partial TFIID complexes. (A) The TFIID-dependent transcription system was supplemented with hTBP-TAFII250 (lanes 1 to 2 and 7 to 8), hTBP-TAFII250-TAFII135 (lanes 3, 4, 9, and 10), and f-TFIID (lanes 5, 6, 11, and 12). Added amounts of TFIID and partial TFIID complexes contained the same amount of TBP (4 ng). Transcription was tested in the absence (lanes 1, 3, 5, 7, 9, and 11) or presence (lanes 2, 4, 6, 8, 10, and 12) of 80 ng of TRα-TRAP complex (12 ng of TRα). Recombinant TFIIA was added to lanes 7 to 12. The TRα-responsive template was TRE3MLΔ53 (50 ng), and the control template consisted of 20 ng of pML200, which contains the major late core promoter. Relative transcriptional activities (quantitated by phosphorimager analysis) from the TR-responsive template were 1 (lanes 1, 5, and 11), 2 (lane 3), 2.4 (lane 7), 4.6 (lane 9), 17 (lane 2), 24 (lane 4), 28 (lanes 8 and 10), 30 (lane 6), and 56 (lane 12). (B) Transcription was performed as in A except that f-TFIID was added to lanes 1, 2, 5, and 6 and TBP was added to lanes 3, 4, 7, and 8. Recombinant TFIIA was added to lanes 5 to 8. Relative transcriptional activities from the TR-responsive template were 1 (lane 3), 1.3 (lanes 1 and 5), 3 (lane 7), 11 (lane 8), 14 (lane 4), 42 (lane 2), and 56 (lane 6). The lower band, below the arrow indicating pML200, is a transcript generated from the activator-responsive template.
FIG. 8
Synergy of TAFIIs with the TRAP complex. The TFIID-dependent transcription system was supplemented with either f-TFIID (lanes 1 to 4) or hTBP (lanes 5 to 8). TRAP complex was added to lanes 3, 4, 7, and 8. Transcription was tested in the absence (lanes 1, 3, 5, and 7) or presence (lanes 2, 4, 6, and 8) of 20 ng of purified fGAL-p65. Added amounts of f-TFIID and TBP alone contained the same amount of TBP (4 ng). The test template contains the adenovirus E1b TATA and initiator regions and consisted of 50 ng of pG5E1b template. The control template consisted of 20 ng of pMLΔ53 and contains the major late core promoter sequence. Relative transcriptional activities from the fGAL4p65-responsive template were: 1 (lanes 5 and 7), 3 (lane 2), 10 (lanes 6 and 8), and 14 (lane 4). Basal levels of transcription with TFIID, either with or without TRAPs (lanes 3 and 1, respectively), were too low to be determined, in contrast to the low but measurable levels of basal transcription with TBP (lanes 5 and 7). The lower band, below the arrow indicating pMLΔ53, is a transcript generated from the activator-responsive template.
Similar articles
- Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.
Salisbury L, Baraitser L. Salisbury L, et al. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review. - Antioxidants for female subfertility.
Showell MG, Mackenzie-Proctor R, Jordan V, Hart RJ. Showell MG, et al. Cochrane Database Syst Rev. 2020 Aug 27;8(8):CD007807. doi: 10.1002/14651858.CD007807.pub4. Cochrane Database Syst Rev. 2020. PMID: 32851663 Free PMC article. - Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.
Lee CC, Chen CW, Yen HK, Lin YP, Lai CY, Wang JL, Groot OQ, Janssen SJ, Schwab JH, Hsu FM, Lin WH. Lee CC, et al. Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924 - Falls prevention interventions for community-dwelling older adults: systematic review and meta-analysis of benefits, harms, and patient values and preferences.
Pillay J, Gaudet LA, Saba S, Vandermeer B, Ashiq AR, Wingert A, Hartling L. Pillay J, et al. Syst Rev. 2024 Nov 26;13(1):289. doi: 10.1186/s13643-024-02681-3. Syst Rev. 2024. PMID: 39593159 Free PMC article. - Antioxidants for female subfertility.
Showell MG, Mackenzie-Proctor R, Jordan V, Hart RJ. Showell MG, et al. Cochrane Database Syst Rev. 2017 Jul 28;7(7):CD007807. doi: 10.1002/14651858.CD007807.pub3. Cochrane Database Syst Rev. 2017. PMID: 28752910 Free PMC article. Updated. Review.
Cited by
- Activation domain-mediator interactions promote transcription preinitiation complex assembly on promoter DNA.
Cantin GT, Stevens JL, Berk AJ. Cantin GT, et al. Proc Natl Acad Sci U S A. 2003 Oct 14;100(21):12003-8. doi: 10.1073/pnas.2035253100. Epub 2003 Sep 23. Proc Natl Acad Sci U S A. 2003. PMID: 14506297 Free PMC article. - The C Terminus of the RNA Polymerase II Transcription Factor IID (TFIID) Subunit Taf2 Mediates Stable Association of Subunit Taf14 into the Yeast TFIID Complex.
Feigerle JT, Weil PA. Feigerle JT, et al. J Biol Chem. 2016 Oct 21;291(43):22721-22740. doi: 10.1074/jbc.M116.751107. Epub 2016 Sep 1. J Biol Chem. 2016. PMID: 27587401 Free PMC article. - Molecular characterization of Saccharomyces cerevisiae TFIID.
Sanders SL, Garbett KA, Weil PA. Sanders SL, et al. Mol Cell Biol. 2002 Aug;22(16):6000-13. doi: 10.1128/MCB.22.16.6000-6013.2002. Mol Cell Biol. 2002. PMID: 12138208 Free PMC article. - Structural changes in TAF4b-TFIID correlate with promoter selectivity.
Liu WL, Coleman RA, Grob P, King DS, Florens L, Washburn MP, Geles KG, Yang JL, Ramey V, Nogales E, Tjian R. Liu WL, et al. Mol Cell. 2008 Jan 18;29(1):81-91. doi: 10.1016/j.molcel.2007.11.003. Mol Cell. 2008. PMID: 18206971 Free PMC article. - Nucleosome remodeling and transcriptional repression are distinct functions of Isw1 in Saccharomyces cerevisiae.
Pinskaya M, Nair A, Clynes D, Morillon A, Mellor J. Pinskaya M, et al. Mol Cell Biol. 2009 May;29(9):2419-30. doi: 10.1128/MCB.01050-08. Epub 2009 Mar 9. Mol Cell Biol. 2009. PMID: 19273607 Free PMC article.
References
- Andel F, III, Ladurner A G, Inouye C, Tjian R, Nogales E. Three-dimensional structure of the human TFIID-IIA-IIB complex. Science. 1999;286:2153–2156. - PubMed
- Birck C, Poch O, Romier C, Ruff M, Mengus G, Lavigne A C, Davidson I, Moras D. Human TAF(II)28 and TAF(II)18 interact through a histone fold encoded by atypical evolutionary conserved motifs also found in the SPT3. Cell. 1998;94:239–249. - PubMed
- Bitter G A. Purification of DNA-dependent RNA polymerase II from Saccharomyces cerevisiae. Anal Biochem. 1983;128:294–310. - PubMed
- Blanar M A, Rutter W J. Interaction cloning: identification of a helix-loop-helix zipper protein that interacts with c-Fos. Science. 1992;256:1014–1018. - PubMed
- Brand M, Leurent C, Mallouh V, Tora L, Schultz P. Three-dimensional structures of the TAFII-containing complexes TFIID and TFTC. Science. 1999;286:2151–2153. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources