Versatile role for hnRNP D isoforms in the differential regulation of cytoplasmic mRNA turnover - PubMed (original) (raw)
Versatile role for hnRNP D isoforms in the differential regulation of cytoplasmic mRNA turnover
N Xu et al. Mol Cell Biol. 2001 Oct.
Abstract
An important emerging theme is that heterogeneous nuclear ribonucleoproteins (hnRNPs) not only function in the nucleus but also control the fates of mRNAs in the cytoplasm. Here, we show that hnRNP D plays a versatile role in cytoplasmic mRNA turnover by functioning as a negative regulator in an isoform-specific and cell-type-dependent manner. We found that hnRNP D discriminates among the three classes of AU-rich elements (AREs), most effectively blocking rapid decay directed by class II AREs found in mRNAs encoding cytokines. Our experiments identified the overlapping AUUUA motifs, one critical characteristic of class II AREs, to be the key feature recognized in vivo by hnRNP D for its negative effect on ARE-mediated mRNA decay. The four hnRNP D isoforms, while differing in their ability to block decay of ARE-containing mRNAs, all potently inhibited mRNA decay directed by another mRNA cis element that shares no sequence similarity with AREs, the purine-rich c-fos protein-coding region determinant of instability. Further experiments indicated that different mechanisms underlie the inhibitory effect of hnRNP D on the two distinct mRNA decay pathways. Our study identifies a potential mechanism by which cytoplasmic mRNA turnover can be differentially and selectively regulated by hnRNP D isoforms in mammalian cells. Our results support the notion that hnRNP D serves as a key factor broadly involved in general mRNA decay.
Figures
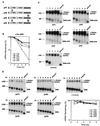
FIG. 1
Ectopic expression of human hnRNP D isoforms in mouse NIH 3T3 cells differentially inhibits rapid mRNA decay directed by the c-fos ARE. (A) Schematic diagram of the four hnRNP D isoforms. The open box indicates the N-terminal domain. RRMI and RRMII depict RNA recognition motifs. The gray box represents the C-terminal domain. The black box at the very N terminus indicates the myc-epitope tag. The striped box represents the additional peptide sequences included as a result of alternative RNA splicing. (B) Semi-log plot showing the effects of four hnRNP D isoforms on mRNA decay directed by the c-fos ARE. Quantitation of mRNA was obtained by scanning radioactive blots with an imager (Packard) and the data were plotted as a function of time. (C) RNA blots showing deadenylation and decay of β-globin mRNA bearing the c-fos ARE (BBB+ARE_fos_) in the absence (control) or presence of ectopically expressed individual isoforms of hnRNP D (indicated by their molecular masses). (D) Northern blots showing decay of β-globin mRNA (BBB) in NIH 3T3 B2A2 cells expressing individual isoforms of hnRNP D or vector only (control). To determine mRNA half-life, NIH 3T3 B2A2 cells were transiently cotransfected with a control plasmid (pSVα-globin/GAPDH) and one of the test plasmids as indicated under each blot. Total cytoplasmic mRNA was isolated at various time intervals after serum stimulation of quiescent cells and analyzed by Northern blot analysis. Transcription of BBB+ARE_fos_ or BBB mRNA was driven by the serum-inducible c-fos promoter. The control mRNA (α-globin/GAPDH) was expressed constitutively and served as an internal standard. The times given at the top correspond to hours after serum stimulation. Poly(A)− RNA was prepared in vitro by treating RNA samples from the 1-h time point with oligo(dT) and RNase H. The positions corresponding to 1,230 and 984 nt are indicated.
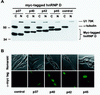
FIG. 2
Expression levels and subcellular distributions of human hnRNP D isoforms in mouse NIH 3T3 cells. (A) Western blot analysis. Cytoplasmic (C) and nuclear (N) lysates prepared from NIH 3T3 B2A2 cells, transfected without (control) or with individual plasmids containing myc-tagged isoforms of hnRNP D, were resolved on a sodium dodecyl sulfate–12% polyacrylamide gel. The blot was probed with a MAb against myc-epitope tag (9E10) (for hnRNP D proteins) and a control MAb against α-tubulin (for cytoplasmic lysate) as well as a control polyclonal antibody against U1 70K (for nuclear lysate). (B) Indirect immunofluorescence microscopy study with the 9E10 MAb showing the subcellular distribution of hnRNP D isoforms (depicted by their molecular masses) and control. Both phase contrast (upper panels) and immunofluorescence (lower panels) views are shown.
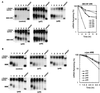
FIG. 3
Ectopic expression of human hnRNP D isoforms in mouse NIH 3T3 cells effectively blocks class II ARE- and not class III ARE-mediated mRNA decay. RNA blots show deadenylation and decay of β-globin mRNA bearing a class II ARE (BBB+AREGM-CSF) (A) or bearing a class III ARE (BBB+AREc-jun) (B) in the absence (control) or presence of ectopically expressed individual isoforms of hnRNP D (indicated by their molecular masses). Semi-log plots show the effects of four hnRNP D isoforms on mRNA decay directed by the GM-CSF ARE (A) or c-jun ARE (B). Cell culture, transfection, gene expression, RNase H treatment, and quantitation of mRNA and data plotting were as described in the legend to Fig. 1.
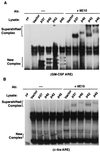
FIG. 4
In vitro interactions of hnRNP D isoforms with different AREs correlate with the copy number of AUUUA motifs in AREs. 32P-labeled RNA transcribed in vitro from human GM-CSF ARE (A) or c-fos ARE (B) was incubated with cytoplasmic lysates from NIH 3T3 B2A2 cells transfected with individual plasmids expressing vector or myc-tagged hnRNP D isoforms. The RNA substrate was incubated with cytoplasmic lysate followed by RNase T1 digestion. The digestion mixtures were left alone (−) or further incubated in the presence of MAb against the myc-epitope tag (+9E10) as indicated on the top of each lane. The final reaction mixtures were analyzed by electrophoresis in a 6% nondenaturing polyacrylamide gel. New complexes (asterisks) and supershifted complexes (brackets) are as indicated.
FIG. 5
hnRNP D isoforms effectively inhibit class II ARE-mediated mRNA decay by recognizing multiple overlapping AUUUA motifs. (A) RNA blots and semi-log plots showing decay of BBB+ARE mRNAs representing individual classes of AREs in NIH 3T3 B2A2 cells expressing p37 isoform (+p37) or vector (control). (B) RNA blots and semi-log plots showing decay of BBB mRNAs containing a mutant GM-CSF ARE (GM1, class I ARE) and a synthetic ARE representing a class II ARE in NIH 3T3 B2A2 cells expressing the p37 isoform of hnRNP D (+p37) or vector (control). Open rectangles and oval symbols in both panels depict AREs and AUUUA motifs, respectively. Cell culture, transfection, RNase H treatment, and quantitation of mRNA and data plotting were as described in the legend to Fig. 1.
FIG. 6
All hnRNP D isoforms effectively block mRNA decay directed by the c-fos coding region instability determinants. (A) RNA blots showing decay of a hybrid mRNA containing the entire c-fos protein-coding region (BFB) in NIH 3T3 B2A2 cells expressing individual isoforms of hnRNP D, HuR (another ARE-BP), or vector (control). Due to comigration of BFB and α-globin control mRNAs, blots were first probed with c-fos coding region probe (for BFB message) and then stripped and reprobed with α-globin-specific probe (for control message). (B) Semi-log plot showing decay of BFB mRNA. Cell culture, transfection, RNase H treatment, and quantitation of mRNA and data plotting were as described in the legend to Fig. 1.
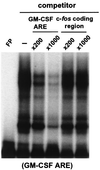
FIG. 7
Ectopically expressed human hnRNP D isoforms do not directly interact with the c-fos coding region in vitro. 32P-labeled RNA transcribed in vitro from human GM-CSF ARE was incubated with cytoplasmic lysates in the presence of increasing amounts of either unlabeled GM-CSF ARE or unlabeled c-fos coding region transcript as competitor. The binding mixtures were analyzed by electrophoresis on a 6% nondenaturing polyacrylamide gel. Gel mobility shift and antibody-supershift assays were carried out as described in the legend to Fig. 4.
Similar articles
- Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element.
Loflin P, Chen CY, Shyu AB. Loflin P, et al. Genes Dev. 1999 Jul 15;13(14):1884-97. doi: 10.1101/gad.13.14.1884. Genes Dev. 1999. PMID: 10421639 Free PMC article. - Distinct RNP complexes of shuttling hnRNP proteins with pre-mRNA and mRNA: candidate intermediates in formation and export of mRNA.
Mili S, Shu HJ, Zhao Y, Piñol-Roma S. Mili S, et al. Mol Cell Biol. 2001 Nov;21(21):7307-19. doi: 10.1128/MCB.21.21.7307-7319.2001. Mol Cell Biol. 2001. PMID: 11585913 Free PMC article. - Highly selective actions of HuR in antagonizing AU-rich element-mediated mRNA destabilization.
Chen CY, Xu N, Shyu AB. Chen CY, et al. Mol Cell Biol. 2002 Oct;22(20):7268-78. doi: 10.1128/MCB.22.20.7268-7278.2002. Mol Cell Biol. 2002. PMID: 12242302 Free PMC article. - Functional diversity of hnRNP proteins.
Singh OP. Singh OP. Indian J Biochem Biophys. 2001 Jun;38(3):129-34. Indian J Biochem Biophys. 2001. PMID: 11693373 Review. - hnRNP complexes: composition, structure, and function.
Krecic AM, Swanson MS. Krecic AM, et al. Curr Opin Cell Biol. 1999 Jun;11(3):363-71. doi: 10.1016/S0955-0674(99)80051-9. Curr Opin Cell Biol. 1999. PMID: 10395553 Review.
Cited by
- A novel role for shuttling SR proteins in mRNA translation.
Sanford JR, Gray NK, Beckmann K, Cáceres JF. Sanford JR, et al. Genes Dev. 2004 Apr 1;18(7):755-68. doi: 10.1101/gad.286404. Genes Dev. 2004. PMID: 15082528 Free PMC article. - cAMP-dependent posttranscriptional regulation of steroidogenic acute regulatory (STAR) protein by the zinc finger protein ZFP36L1/TIS11b.
Duan H, Cherradi N, Feige JJ, Jefcoate C. Duan H, et al. Mol Endocrinol. 2009 Apr;23(4):497-509. doi: 10.1210/me.2008-0296. Epub 2009 Jan 29. Mol Endocrinol. 2009. PMID: 19179481 Free PMC article. - DEAD box protein DDX1 regulates cytoplasmic localization of KSRP.
Chou CF, Lin WJ, Lin CC, Luber CA, Godbout R, Mann M, Chen CY. Chou CF, et al. PLoS One. 2013 Sep 4;8(9):e73752. doi: 10.1371/journal.pone.0073752. eCollection 2013. PLoS One. 2013. PMID: 24023901 Free PMC article. - Post-transcriptional control of gene expression by AUF1: mechanisms, physiological targets, and regulation.
White EJ, Brewer G, Wilson GM. White EJ, et al. Biochim Biophys Acta. 2013 Jun-Jul;1829(6-7):680-8. doi: 10.1016/j.bbagrm.2012.12.002. Epub 2012 Dec 14. Biochim Biophys Acta. 2013. PMID: 23246978 Free PMC article. Review. - Destabilization of interleukin-6 mRNA requires a putative RNA stem-loop structure, an AU-rich element, and the RNA-binding protein AUF1.
Paschoud S, Dogar AM, Kuntz C, Grisoni-Neupert B, Richman L, Kühn LC. Paschoud S, et al. Mol Cell Biol. 2006 Nov;26(22):8228-41. doi: 10.1128/MCB.01155-06. Epub 2006 Sep 5. Mol Cell Biol. 2006. PMID: 16954375 Free PMC article.
References
- Chen A C-Y, Shyu A-B. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources