Novel meiosis-specific isoform of mammalian SMC1 - PubMed (original) (raw)
Novel meiosis-specific isoform of mammalian SMC1
E Revenkova et al. Mol Cell Biol. 2001 Oct.
Abstract
Structural maintenance of chromosomes (SMC) proteins fulfill pivotal roles in chromosome dynamics. In yeast, the SMC1-SMC3 heterodimer is required for meiotic sister chromatid cohesion and DNA recombination. Little is known, however, about mammalian SMC proteins in meiotic cells. We have identified a novel SMC protein (SMC1beta), which-except for a unique, basic, DNA binding C-terminal motif-is highly homologous to SMC1 (which may now be called SMC1alpha) and is not present in the yeast genome. SMC1beta is specifically expressed in testes and coimmunoprecipitates with SMC3 from testis nuclear extracts, but not from a variety of somatic cells. This establishes for mammalian cells the concept of cell-type- and tissue-specific SMC protein isoforms. Analysis of testis sections and chromosome spreads of various stages of meiosis revealed localization of SMC1beta along the axial elements of synaptonemal complexes in prophase I. Most SMC1beta dissociates from the chromosome arms in late-pachytene-diplotene cells. However, SMC1beta, but not SMC1alpha, remains chromatin associated at the centromeres up to metaphase II. Thus, SMC1beta and not SMC1alpha is likely involved in maintaining cohesion between sister centromeres until anaphase II.
Figures
FIG. 1
Immunoprecipitation of SMC3 and associated SMC1 proteins with anti-SMC3 antibodies. (A) Immunoprecipitation from various bovine tissue nuclear extracts. Thym., thymus. (B) Immunoprecipitation from mouse and bovine testis nuclear extracts. (C) Control immunoprecipitation from bovine testis nuclear extract, with (+Ab) and without (−Ab) anti-SMC3 antibody included. (D) Immunoprecipitation from nuclear extracts prepared from actively proliferating (lipopolysaccharide-induced; Activ.) and resting (Restg.) mouse spleen cell cultures and from an actively growing mouse pre-B-cell line. (E) Immunoprecipitation from bovine testis nuclear extract gel filtration (BioGel A15m resin) fractions (20 μg of protein each) representing 1-, 3-, and 6-MDa molecular-mass positions. (F) Immunoprecipitation from mouse kidney, rat testis, and purified rat spermatocyte (Purif. Sp.) nuclear extracts (25 μg of protein each). Controls without antibody (no Ab) are included. All precipitates were analyzed by SDS-polyacrylamide gel electrophoresis and silver staining. M, molecular mass marker.
FIG. 2
Amino acid sequence comparison and dendrogram of SMC1β. (A) N-terminal domains of SMC1β and mammalian SMC proteins representing the four most closely related SMC subfamilies. (B) C-terminal domains of SMC1β and mammalian SMC proteins representing the most closely related SMC subfamilies. The program Megalign (DNAStar) was used. The accession numbers for the related SMC proteins are as follows: mSMC1α (mouse SMCB; AF047600), bSMC1α (bovine SMC1; AF072712), hSMC1α (human SB1.8; S78271), XSMC1 (X. laevis; AF051784), mSMC3 (mouse SMCD; AF047601), bSMC3 (bovine; AF072713), hSMC4 (human CAP-C; AB019987), and hSMC2 (human CAP-E; AF092563). mSMC1β, mouse SMC1β; hSMC1β, predicted human protein (CAB41703; partial sequence). Identical residues are shaded. Dashes indicate gaps in alignment.
FIG. 2
Amino acid sequence comparison and dendrogram of SMC1β. (A) N-terminal domains of SMC1β and mammalian SMC proteins representing the four most closely related SMC subfamilies. (B) C-terminal domains of SMC1β and mammalian SMC proteins representing the most closely related SMC subfamilies. The program Megalign (DNAStar) was used. The accession numbers for the related SMC proteins are as follows: mSMC1α (mouse SMCB; AF047600), bSMC1α (bovine SMC1; AF072712), hSMC1α (human SB1.8; S78271), XSMC1 (X. laevis; AF051784), mSMC3 (mouse SMCD; AF047601), bSMC3 (bovine; AF072713), hSMC4 (human CAP-C; AB019987), and hSMC2 (human CAP-E; AF092563). mSMC1β, mouse SMC1β; hSMC1β, predicted human protein (CAB41703; partial sequence). Identical residues are shaded. Dashes indicate gaps in alignment.
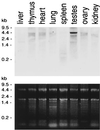
FIG. 3
Tissue specificity of SMC1β RNA expression. (Top) Northern blot of total RNA extracted from different mouse tissues and hybridized to an SMC1β-specific probe. (Bottom) Corresponding agarose gel stained with Radiant Red fluorescent RNA stain prior to transfer for loading control.
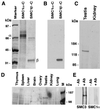
FIG. 4
Generation and specificity of anti-SMC1β monoclonal antibodies. (A) Coomassie blue-stained SDS-polyacrylamide gel loaded with the C-terminal domains of SMC1α (approximately 67 kDa) and SMC1β (approximately 33 kDa), with their positions indicated by α and β. (B) Immunoblot of a gel identical to that in panel A probed with a monoclonal anti-SMC1β antibody. (C) Immunoblot of nuclear extracts from mouse testis and kidney probed with a monoclonal anti-SMC1β antibody. (D) Immunoblot of nuclear extracts from a variety of bovine tissues probed with a monoclonal anti-SMC1β antibody. M, marker. (E) Anti-SMC1β immunoprecipitates from testis nuclear extracts probed in Western blotting with anti-SMC3 or anti-SMC1α antibodies. −AB, no anti-SMC1β; +AB, with anti-SMC1β.
FIG. 5
Testis and liver section stained with anti-SMC1β. Sections of mouse testis or liver were incubated with either propidium iodide (PI)-RNase or anti-SMC1β, FITC-labeled, to visualize DNA or SMC1β. The merged images are at the bottom, and two magnifications are shown as indicated. Bar = 10 μm.
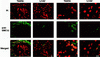
FIG. 6
Immunolocalization of SMC1β and SMC1α in successive stages of meiosis. (A to J) Images of immunofluorescence triple labeling of SCP3 (blue), SMC1β (green), and kinetochores (red) in dry-down preparations of rat spermatocytes. (A to F and H) Shown on the left are the merged images of SCP3 (blue) and kinetochores (red), and shown on the right are the merged images of the same cell of SMC1β (green) and kinetochores (red). (A) zygotene (the long arrows indicate asynapsed segments of AEs, the short arrows point to regions of presynaptic alignment, and the arrowheads designate paired segments of AEs); (B) pachytene (the XY bivalent is indicated by arrows); (C) diplotene (the arrows point to connections containing SCP3 and SMC1β between AEs of homologous chromosomes); (D) diakinesis (the arrowheads point to partial splitting of AEs); (E) metaphase I (the arrowheads indicate a weak signal for SCP3 in the chromosome arms, whereas SMC1β is hardly detectable); (F) metaphase II; (H) anaphase II. (G) Enlargement of the area indicated in panel F, with the merged images of SCP3 (blue) and kinetochores (red) (left); SMC1β (green) and kinetochores (red) (middle); and SCP3 (blue), SMC1β (green), and kinetochores (red) (right). (I and J) Enlargements of areas indicated in panel H, with the merged images of SCP3 (blue) and kinetochores (red) (left); SMC1β (green) and kinetochores (red) (middle); and SCP3 (blue), SMC1β (green), and kinetochores (red) (right). (K to M) Images of immunofluorescence triple labeling of SCP3 (blue), SMC1β (green), and SMC1α (red). Shown are a pachytene SC (K), a diplotene SC (L), and a metaphase I bivalent (M) in a dry-down preparation of rat spermatocytes. The tops of panels K to M show the merged images of SCP3 (blue), SMC1α (red), and SMC1β (green); the middles show SCP3 (blue), SMC1β (green), and SMC1α (red); and the bottoms show SCP3 (blue), SMC1β (green), and SMC1α (red). Bars = 10 μm (A to F and H) and 1 μm (G, I, J, and K to M).
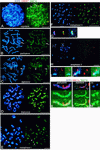
FIG. 7
Immunolocalization of SMC3 and SMC1α in successive stages of meiosis. (A to C) Images of immunofluorescence triple labeling of SMC1α (red), SMC3 (green), and SCP3 (blue) in a frozen section of rat testis. (A) Merged images of SMC1α (red) and SMC3 (green). (B) Merged images of SCP3 (blue) and SMC1α (red). (C) Merged images of SMC1α (red), SMC3 (green), and SCP3 (blue). The pictures show parts of two testicular tubules. lp, late-pachytene spermatocytes; pl, preleptotene spermatocytes; i, interstitial zone between the two tubules; lz, late-zygotene spermatocytes. (D) Images of immunofluorescence triple labeling of SMC1α (red), SMC3 (green), and SCP3 (blue) in a rat pachytene nucleus, spread by agar filtration; on the left, the merged images of SCP3 (blue) and SMC1α (red) are shown, and on the right, the merged images of SMC3 (green) and SMC1α (red) are shown. (E to K) Images of immunofluorescence triple labeling of SCP3 (blue), SMC3 (green), and kinetochores (red) in dry-down preparations of rat spermatocytes. (E to I) On the left are the merged images of SCP3 (blue) and kinetochores (red), and on the right are SMC3 (green) and kinetochores (red). (J) Enlarged images of the area indicated in H. (K) Enlarged images of the area indicated in I. On the left are the merged images of SCP3 (blue) and kinetochores (red), in the middle are SMC3 (green) and kinetochores (red), and on the right are SCP3 (blue), SMC3 (green), and kinetochores (red). Bars = 10 μm (A to I) and 1 μm (J and K).
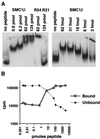
FIG. 8
DNA interaction of the 28-aa SMC1β C-terminal motif. (A) In a gel shift assay, increasing amounts of the 28-aa peptide were incubated with 0.8 pmol of a 5′-32P-labeled 200-bp ribosomal DNA fragment as indicated. R54 and R21, Rad54 and Rad21 control peptides. (B) Assay for protein-DNA network formation. Bound (pelleted) and unbound (supernatant) DNA was measured after incubation of 1.5-pmol of DNA substrate with increasing amounts of the 28-aa peptide.
Similar articles
- Temporally and spatially selective loss of Rec8 protein from meiotic chromosomes during mammalian meiosis.
Lee J, Iwai T, Yokota T, Yamashita M. Lee J, et al. J Cell Sci. 2003 Jul 1;116(Pt 13):2781-90. doi: 10.1242/jcs.00495. Epub 2003 May 20. J Cell Sci. 2003. PMID: 12759374 - Meiotic cohesin REC8 marks the axial elements of rat synaptonemal complexes before cohesins SMC1beta and SMC3.
Eijpe M, Offenberg H, Jessberger R, Revenkova E, Heyting C. Eijpe M, et al. J Cell Biol. 2003 Mar 3;160(5):657-70. doi: 10.1083/jcb.200212080. J Cell Biol. 2003. PMID: 12615909 Free PMC article. - Changes in the expression and localization of cohesin subunits during meiosis in a non-mammalian vertebrate, the medaka fish.
Iwai T, Lee J, Yoshii A, Yokota T, Mita K, Yamashita M. Iwai T, et al. Gene Expr Patterns. 2004 Sep;4(5):495-504. doi: 10.1016/j.modgep.2004.03.004. Gene Expr Patterns. 2004. PMID: 15261826 - The many functions of SMC proteins in chromosome dynamics.
Jessberger R. Jessberger R. Nat Rev Mol Cell Biol. 2002 Oct;3(10):767-78. doi: 10.1038/nrm930. Nat Rev Mol Cell Biol. 2002. PMID: 12360193 Review. - Meiotic prophase-like pathway for cleavage-independent removal of cohesin for chromosome morphogenesis.
Challa K, Shinohara M, Shinohara A. Challa K, et al. Curr Genet. 2019 Aug;65(4):817-827. doi: 10.1007/s00294-019-00959-x. Epub 2019 Mar 28. Curr Genet. 2019. PMID: 30923890 Review.
Cited by
- Cohesin SMC1beta protects telomeres in meiocytes.
Adelfalk C, Janschek J, Revenkova E, Blei C, Liebe B, Göb E, Alsheimer M, Benavente R, de Boer E, Novak I, Höög C, Scherthan H, Jessberger R. Adelfalk C, et al. J Cell Biol. 2009 Oct 19;187(2):185-99. doi: 10.1083/jcb.200808016. J Cell Biol. 2009. PMID: 19841137 Free PMC article. - Meiotic sex chromosome cohesion and autosomal synapsis are supported by Esco2.
McNicoll F, Kühnel A, Biswas U, Hempel K, Whelan G, Eichele G, Jessberger R. McNicoll F, et al. Life Sci Alliance. 2020 Feb 12;3(3):e201900564. doi: 10.26508/lsa.201900564. Print 2020 Mar. Life Sci Alliance. 2020. PMID: 32051254 Free PMC article. - Chromosome Organization in Early Meiotic Prophase.
Grey C, de Massy B. Grey C, et al. Front Cell Dev Biol. 2021 Jun 3;9:688878. doi: 10.3389/fcell.2021.688878. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34150782 Free PMC article. Review. - Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA-ATPase.
Wojtasz L, Daniel K, Roig I, Bolcun-Filas E, Xu H, Boonsanay V, Eckmann CR, Cooke HJ, Jasin M, Keeney S, McKay MJ, Toth A. Wojtasz L, et al. PLoS Genet. 2009 Oct;5(10):e1000702. doi: 10.1371/journal.pgen.1000702. Epub 2009 Oct 23. PLoS Genet. 2009. PMID: 19851446 Free PMC article. - Structural maintenance of chromosomes (SMC) proteins, a family of conserved ATPases.
Harvey SH, Krien MJ, O'Connell MJ. Harvey SH, et al. Genome Biol. 2002;3(2):REVIEWS3003. doi: 10.1186/gb-2002-3-2-reviews3003. Epub 2002 Jan 30. Genome Biol. 2002. PMID: 11864377 Free PMC article. Review.
References
- Akhmedov A T, Frei M, Tsai-Pflugfelder C, Kemper B, Gasser S M, Jessberger R. Structural maintenance of chromosomes: protein C-terminal domains bind preferentially to DNA with secondary structure. J Biol Chem. 1998;273:24088–24094. - PubMed
- Akhmedov A T, Gross B, Jessberger R. Mammalian SMC3 C-terminal and coiled-coil protein domains specifically bind palindromic DNA, do not block DNA ends, and prevent DNA bending. J Biol Chem. 1999;274:38216–38224. - PubMed
- Bayne M L, Alexander R F, Benbow R M. DNA binding protein from ovaries of the frog Xenopus laevis which promotes concatenation of linear DNA. J Mol Biol. 1984;172:87–108. - PubMed
- Bishop D K, Park D, Xu L, Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous