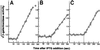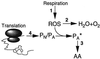Bacterial senescence: protein oxidation in non-proliferating cells is dictated by the accuracy of the ribosomes - PubMed (original) (raw)
Bacterial senescence: protein oxidation in non-proliferating cells is dictated by the accuracy of the ribosomes
M Ballesteros et al. EMBO J. 2001.
Abstract
We have investigated the causal factors behind the age-related oxidation of proteins during arrest of cell proliferation. A proteomic approach demonstrated that protein oxidation in non-proliferating cells is observed primarily for proteins being produced in a number of aberrant isoforms. Also, these cells exhibited a reduced translational fidelity as demonstrated by both proteomic analysis and genetic measurements of nonsense suppression. Mutants harboring hyperaccurate ribosomes exhibited a drastically attenuated protein oxidation during growth arrest. In contrast, oxidation was augmented in mutants with error-prone ribosomes. Oxidation increased concomitantly with a reduced rate of translation, indicating that the production of aberrant, and oxidized proteins, is not the result of titration of the co-translational folding machinery. The age-related accumulation of the chaperones, DnaK and GroEL, was drastically attenuated in the hyperaccurate rpsL mutant, demonstrating that the reduced translational fidelity in growth-arrested cells may also be a primary cause for the induction of the heat shock regulon. The data point to an alternative way of approaching the causal factors involved in protein oxidation in eukaryotic G(0) cells.
Figures
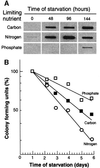
Fig. 1. (A) Protein oxidation in non-proliferating cells arrested by starvation for different nutrients. The autoradiogram shows carbonyl contents in wild-type E.coli cells during starvation for carbon, nitrogen or phosphate (as indicated). Equal amounts of protein (1 µg) were loaded in each slot and analysis was repeated three times to confirm reproducibility. Time zero denotes the sample obtained from exponentially growing cells. (B) Viability, measured as colony-forming units, during starvation for different nutrients. The highest colony count obtained immediately after the nutrients were depleted was assigned a value of 100%.
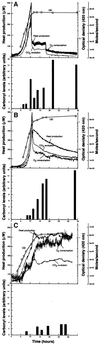
Fig. 2. Correlation between metabolic activity and protein oxidation. Protein carbonyl levels and metabolic activity were analyzed during growth and starvation for carbon (A), nitrogen (B) or phosphate (C). The upper panel in each graph shows growth (measured as OD at 420 nm), heat production, carbon dioxide production and oxygen consumption. The lower panels show protein carbonyl levels quantified using the NIH 1.62 software. All carbonyl values were related to that obtained during exponential growth, which was assigned a value of 1.0.
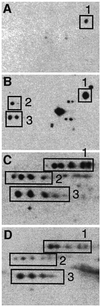
Fig. 3. Correlation between production of aberrant protein isoforms and carbonylation. The pattern of protein carbonylation was determined by two-dimensional western blot immunoassay. The films were obtained after carbonyl immunoassay of exponentially growing wild-type cells (A), exponentially growing wild-type cells exposed to 200 µM H2O2 (B), exponentially growing rpsD mutant (C) and 1 h carbon-starved wild-type cells (D). The autoradiogram depicts the area around the oxidation-sensitive enzyme glutamine synthetase (GlnA; boxed protein no. 1) since three proteins in this area exhibit extensive stuttering.
Fig. 4. Mistranslation in starved, non-proliferating cells. Two-dimensional autoradiograms of (A) growing and (B) starving (1 h glucose starvation) cells obtained after [35S]methionine pulse labeling. The circles show elongation factor, EF-Tu, whereas the heat shock proteins DnaK, GroEL and HtpG are marked with white lines. The boxes mark areas of intense protein stuttering (production of multiple protein isoforms). (C) Protein stuttering in wild-type (upper panel) compared with the rpsL mutant (middle panel). The lower panel shows the V8 protease peptide fingerprints (see Materials and methods) of the protein spots indicated and numbered in the upper panel. (D) In vivo read-through of a nonsense codon in wild-type cells during carbon starvation. The assay for calculating the levels of nonsense suppression is described in Materials and methods. A value of 0.01 indicates that one out of 100 transcripts generates a full-length protein due to nonsense read-through.
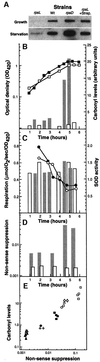
Fig. 5. Effects of translational errors on protein oxidation. (A) Levels of carbonylation during growth and starvation (1 h carbon starved) in wild-type, hyperaccurate (rpsL), with and without 1 µg/ml streptomycin, and sloppy (rpsD) mutants. (B) Carbonylation, respiration, superoxide dismutase activity and nonsense suppression in the rpsL mutant are compared with the wild-type strain. Protein carbonyl contents (bars) and growth (squares) in wild-type (gray bars, closed squares) and the rpsL mutant (open bars, open squares) during growth and entry into stationary phase (glucose starvation). Protein carbonyls were quantified from the autoradiograms using the NIH 1.62 software. (C) Oxygen consumption (circles) and SOD activity (bars) for wild-type (closed circles, gray bars) and rpsL mutant (open circles, open bars) were determined as described in Materials and methods. (D) The frequency of read-through of a nonsense codon in the wild-type (gray bars) and the rpsL mutant (open bars). (E) Correlation between nonsense suppression and protein carbonylation. The data shown are for wild-type (circles), rpsL mutant (triangles) and rpsD mutants (squares) obtained during growth (closed symbols) and carbon starvation (open symbols). Open diamonds and crosses represent values for nitrogen- and phosphate-starved wild-type cells, respectively. In the rpsL mutant, no increase in nonsense suppression was observed in stationary phase. All carbonylation values were compared with the level of the wild-type strain growing exponentially in glucose minimal M9 medium, which was assigned a value of 1.0.
Fig. 6. Determination of peptide chain elongation rates as measured by induction kinetics of β-galactosidase during exponential growth (A) and at 15 (B) and 60 min (C) of glucose starvation. The lac operon was induced by addition of IPTG and, at frequent intervals thereafter, samples were withdrawn and measured for β-galactosidase activity as described in Materials and methods.
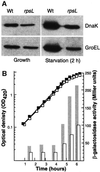
Fig. 7. Effects of translational accuracy on the induction of heat shock proteins during starvation. (A) Analysis of GroEL and DnaK levels in exponentially growing and starving (glucose starvation, 2 h) wild-type and rpsL mutant cells. (B) GroEL promoter activity (bars) during growth and starvation (OD; squares) in wild-type (closed squares, gray bars) and rpsL mutant (open squares, open bars).
Fig. 8. Schematic representation of activities suggested to be involved in causing increased oxidation during aging of cells. Activity 1 is the respiratory activity generating reactive oxygen species (ROS), whereas activity 2 denotes the oxidation defense system, including the superoxide dismutases, catalases and peroxidases. Activity 3 denotes the proteolytic apparatus responsible for degrading oxidation-damaged proteins and peptides, and activity 4 the production of aberrant (PA) and native (PN) proteins by the translation process. PA* denotes aberrant and oxidized proteins and AA amino acids. See text for details.
Similar articles
- Induction of the heat shock regulon in response to increased mistranslation requires oxidative modification of the malformed proteins.
Fredriksson A, Ballesteros M, Dukan S, Nyström T. Fredriksson A, et al. Mol Microbiol. 2006 Jan;59(1):350-9. doi: 10.1111/j.1365-2958.2005.04947.x. Mol Microbiol. 2006. PMID: 16359340 - Protein oxidation in response to increased transcriptional or translational errors.
Dukan S, Farewell A, Ballesteros M, Taddei F, Radman M, Nyström T. Dukan S, et al. Proc Natl Acad Sci U S A. 2000 May 23;97(11):5746-9. doi: 10.1073/pnas.100422497. Proc Natl Acad Sci U S A. 2000. PMID: 10811907 Free PMC article. - Development of a chaperone-deficient system by fractionation of a prokaryotic coupled transcription/translation system.
Kudlicki W, Mouat M, Walterscheid JP, Kramer G, Hardesty B. Kudlicki W, et al. Anal Biochem. 1994 Feb 15;217(1):12-9. doi: 10.1006/abio.1994.1077. Anal Biochem. 1994. PMID: 7911283 - Translational fidelity, protein oxidation, and senescence: lessons from bacteria.
Nyström T. Nyström T. Ageing Res Rev. 2002 Sep;1(4):693-703. doi: 10.1016/s1568-1637(02)00028-4. Ageing Res Rev. 2002. PMID: 12208238 Review. - The translational machinery is an optimized molecular network that affects cellular homoeostasis and disease.
Kazana E, von der Haar T. Kazana E, et al. Biochem Soc Trans. 2014 Feb;42(1):173-6. doi: 10.1042/BST20130131. Biochem Soc Trans. 2014. PMID: 24450647 Review.
Cited by
- Accumulation of oxidized proteins in Herpesvirus infected cells.
Mathew SS, Bryant PW, Burch AD. Mathew SS, et al. Free Radic Biol Med. 2010 Aug 1;49(3):383-91. doi: 10.1016/j.freeradbiomed.2010.04.026. Epub 2010 May 2. Free Radic Biol Med. 2010. PMID: 20441790 Free PMC article. - Starvation for different nutrients in Escherichia coli results in differential modulation of RpoS levels and stability.
Mandel MJ, Silhavy TJ. Mandel MJ, et al. J Bacteriol. 2005 Jan;187(2):434-42. doi: 10.1128/JB.187.2.434-442.2005. J Bacteriol. 2005. PMID: 15629914 Free PMC article. - Errors during Gene Expression: Single-Cell Heterogeneity, Stress Resistance, and Microbe-Host Interactions.
Evans CR, Fan Y, Weiss K, Ling J. Evans CR, et al. mBio. 2018 Jul 3;9(4):e01018-18. doi: 10.1128/mBio.01018-18. mBio. 2018. PMID: 29970467 Free PMC article. Review. - Protein mistranslation protects bacteria against oxidative stress.
Fan Y, Wu J, Ung MH, De Lay N, Cheng C, Ling J. Fan Y, et al. Nucleic Acids Res. 2015 Feb 18;43(3):1740-8. doi: 10.1093/nar/gku1404. Epub 2015 Jan 10. Nucleic Acids Res. 2015. PMID: 25578967 Free PMC article. - Stressed mycobacteria use the chaperone ClpB to sequester irreversibly oxidized proteins asymmetrically within and between cells.
Vaubourgeix J, Lin G, Dhar N, Chenouard N, Jiang X, Botella H, Lupoli T, Mariani O, Yang G, Ouerfelli O, Unser M, Schnappinger D, McKinney J, Nathan C. Vaubourgeix J, et al. Cell Host Microbe. 2015 Feb 11;17(2):178-90. doi: 10.1016/j.chom.2014.12.008. Epub 2015 Jan 22. Cell Host Microbe. 2015. PMID: 25620549 Free PMC article.
References
- Andersson D.I., Bohman,K., Isaksson,L.A. and Kurland,C.G. (1982) Translation rates and misreading characteristics of rpsD mutants in Escherichia coli. Mol. Gen. Genet., 187, 467–472. - PubMed
- Barak Z., Gallant,J., Lindsley,D., Kwieciszewki,B. and Heidel,D. (1996) Enhanced ribosome frameshifting in stationary phase cells. J. Mol. Biol., 263, 140–148. - PubMed
- Beckman K.B. and Ames,B.N. (1998) The free radical theory of aging matures. Physiol. Rev., 78, 547–581. - PubMed
- Benov L. and Fridovich,I. (1995) A superoxide dismutase mimic protects sodA sodB Escherichia coli against aerobic heating and stationary-phase death. Arch. Biochem. Biophys., 322, 291–294. - PubMed
- Berlett B.S. and Stadtman,E.R. (1997) Protein oxidation in aging, disease and oxidative stress. J. Biol. Chem., 272, 20313–20316. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials