A role for cofilin and LIM kinase in Listeria-induced phagocytosis - PubMed (original) (raw)
A role for cofilin and LIM kinase in Listeria-induced phagocytosis
H Bierne et al. J Cell Biol. 2001.
Abstract
The pathogenic bacterium Listeria monocytogenes is able to invade nonphagocytic cells, an essential feature for its pathogenicity. This induced phagocytosis process requires tightly regulated steps of actin polymerization and depolymerization. Here, we investigated how interactions of the invasion protein InlB with mammalian cells control the cytoskeleton during Listeria internalization. By fluorescence microscopy and transfection experiments, we show that the actin-nucleating Arp2/3 complex, the GTPase Rac, LIM kinase (LIMK), and cofilin are key proteins in InlB-induced phagocytosis. Overexpression of LIMK1, which has been shown to phosphorylate and inactivate cofilin, induces accumulation of F-actin beneath entering particles and inhibits internalization. Conversely, inhibition of LIMK's activity by expressing a dominant negative construct, LIMK1(-), or expression of the constitutively active S3A cofilin mutant induces loss of actin filaments at the phagocytic cup and also inhibits phagocytosis. Interestingly, those constructs similarly affect other actin-based phenomenons, such as InlB-induced membrane ruffling or Listeria comet tail formations. Thus, our data provide evidence for a control of phagocytosis by both activation and deactivation of cofilin. We propose a model in which cofilin is involved in the formation and disruption of the phagocytic cup as a result of its local progressive enrichment.
Figures
Figure 1.
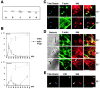
Actin and Met recruitment during InlB-mediated internalization. (A) Diagram of the internalization process: adherent bacteria (a), formation of the phagocytic cup with actin polymerization (b), phagocytic cup closure (c), disruption of the F-actin cup with residual (d) or no (e) F-actin around the phagosome. (B) Kinetic analysis of F-actin cup formation during InlB-mediated entry. Internalized and F-actin–associated InlB particles were quantified by immunofluorescence at different times after infection. Results are expressed as the percentage of intracellular particles (red dashed line) or the percentage of particles associated with an F-actin ring (black solid line) among the total number of cell-associated particles. (C–E) Met recruitment during InlB bead or bacteria entry at different times after infection. (C) Vero cells were stained with FITC-phalloidin (green) and anti- Met Ab (red). InlB beads are intrinsically fluorescent (blue). (D) Total bacteria associated with cells were visualized in phase–contrast, and bacteria that are fully or partly extracellular were detected by labeling with anti-InlA Ab before cell permeabilization (last panel, blue). (E) Extracellular beads were labeled with anti-InlB Ab (green) before cell permeabilization. Arrows indicate intracellular InlB beads or bacteria not associated with Met. Bars, 5 μm.
Figure 2.
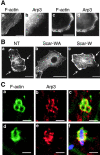
A role for the Arp2/3 complex in InlB-induced membrane ruffles and phagocytosis. (A) Recruitment of the Arp2/3 complex to InlB-induced ruffles. Vero cells, untreated (a and b) or stimulated with 4.5 nM of InlB (c and d) stained with FITC-phalloidin and anti-Arp3 Ab. (B) Inhibition of InlB-induced ruffles in ScarWA- transfected cells. Vero cells, nontransfected (NT, a) or transiently transfected with ScarWA (b) or ScarW (c) stimulated with InlB and stained with FITC-phalloidin. (C) Recruitment of the Arp2/3 complex to InlB beads (a–c) or bacteria- (d–f) induced phagocytic cups. Cells were stained with FITC-phalloidin (green) and anti-Arp3 Ab (red). The bacterium is labeled with anti-InlA Ab (f, blue). Images represent confocal slices of 0.6 (beads) and 1.2 μm (bacteria). Bars: (A) 10 μm; (B) 20 μm; (C) 2 μm.
Figure 3.
Cofilin is recruited to InlB-induced membrane ruffles and phagocytic cups. (A) Colocalization of cofilin with F-actin in InlB-induced ruffles. Vero cells, untreated (a and b) or stimulated with 4.5 nM of InlB (c and d) stained with FITC-phalloidin and anti-cofilin Ab. (B) Recruitment of cofilin by bacteria at different stages of the internalization process in Ref52 cells. The left panels schematically represent the bacteria detected in the five other panels and marked by reference to Fig. 1 A. Total bacteria associated with cells was visualized in phase–contrast, and bacteria that are fully or partly extracellular were identified by labeling with anti-InlA Ab before cell permeabilization. Cells were stained with FITC-phalloidin and anti-cofilin Ab. In the merged images, extracellular bacteria are blue, F-actin is green, and cofilin is red. Arrowheads or arrows indicate bacteria that colocalizes or not with cofilin, respectively. Bars: (A, a) 10 μm; (A, c) 2 μm.
Figure 4.
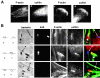
LIMK1 and LIMK1 − expression affect Listeria actin-based motility and InlB-mediated ruffling. Vero cells nontransfected (NT) or transiently transfected with LIMK1 or LIMK1− stained with FITC-phalloidin and with anti-Myc Ab to detect the LIMK1 and LIMK1− fusion proteins. (A) Resting cells. (B) Cells infected with the Listeria variant EGDΔ_inlB_(LRRs-IR-SPA) and analyzed for actin tail formation. F-actin is in green and bacteria are in red. LIMK1 accumulates in long comet tails (arrows). (C) Cells stimulated with 4.5 nM of InlB for 5 min and analyzed for membrane ruffle formation. LIMK1 accumulates in the ruffle areas (arrows). LIMK1(−), LIMK1(−); LIMK1(HT), LIMK1 highly transfected cells. Bars: (A) 20 μm; (B) 10 μm; (C) 20 μm.
Figure 5.
Effects of LIMK1 and LIMK1 − expression on the formation of F-actin cups at the entry site of InlB beads and bacteria. Vero cells nontransfected (NT) or transiently transfected with LIMK1 or LIMK1− incubated with InlB beads for 10 min (A) or with bacteria for 20 min (B) stained with anti-Myc Ab to detect the LIMK1 and LIMK1− fusion proteins and FITC-phalloidin. Boxed regions in the left panels indicate the position of the field, which is magnified in the images on the right panels. In the merged images, total InlB beads are blue, F-actin is green, Myc is red, and the bacteria are visualized in phase–contrast. LIMK1 colocalized with F-actin in abnormal phagocytic cups (arrows). Bars, 5 μm.
Figure 6.
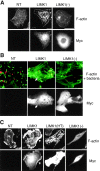
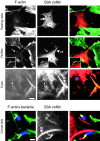
S3A cofilin recruitment to InlB-induced ruffles, InlB-mediated phagocytic cups, and Listeria comet tails decreases the amount of F-actin in these structures. Vero cells transiently transfected with S3A cofilin were left unstimulated (resting cells) or analyzed for the formation of InlB-induced ruffles, phagocytic cups, or Listeria comet tails as described in the legends to Figs. 4 and 5. Cells were stained with FITC-phalloidin, anti-Myc Ab to detect S3 cofilin, and for comet analysis with anti-Listeria Ab. In the merged images, F-actin is green, S3A cofilin is red, and InlB beads or bacteria are blue. S3A cofilin is localized in reduced membrane ruffles (magnified in boxed regions), thin phagocytic cups, and short comet tails (arrows). Arrowheads indicate cofilin rods (rd). Bar: (first and second rows) 10 μm; (boxed images and third and fourth rows) 2 μm.
Figure 7.
In LIMK1-overexpressing cells, endogenous cofilin is still recruited to actin-based structures. Vero cells transiently transfected with LIMK1 were analyzed for Listeria tail formation, InlB-induced ruffling, or bacterial entry as described in the legends to Figs. 4 and 5. Cells were stained with FITC-phalloidin, anti-Myc, and anti- cofilin Ab. In the merged images, cofilin is green and LIMK1 is red. Bars: (first and second rows) 10 μm; (third row) 2 μm.
Figure 8.
Overexpression of S3A cofilin suppresses F-actin accumulation induced by LIMK1 overexpression. Vero cells transiently cotransfected with LIMK1 and S3A cofilin cDNAs were analyzed for formation of Listeria tails, InlB-induced ruffles, or InlB bead phagocytic cups as described in the legends to Figs. 4 and 5. Cells were stained with FITC-phalloidin, anti-Myc, which detects both LIMK1 and S3A fusion proteins and lights up cofilin rods (arrowheads, rd), and an antipeptide Ab specific only to the LIMK1 fusion protein. For cups, boxed regions indicate the position of the field, which is magnified below. Due to their intrinsic fluorescence in the 650–700-nM range, InlB beads are detected together with the Cy5-labeled LIMK1 anti-tag. In the merged images, F-actin is green, Myc is red, and LIMK1 and InlB beads are blue. Bars: (full images) 10 μm; (magnified images) 2 μm.
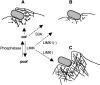
Figure 9.
Possible role of the cofilin phosphocycle in InlB- induced phagocytosis. (A) Interaction of InlB with its receptors triggers actin polymerization and formation of the phagocytic cup. This step involves recruitment of (1) the Arp2/3 complex (open circles), which promotes actin nucleation and branching of the filaments, and (2) ADF/cofilin (closed circles), which stimulates actin dynamics by severing actin filaments, creating new ends that allow actin polymerization, and by depolymerizing actin filaments, supplying new actin monomers. LIMK recruited at this step could prevent excessive depolymerization of the filaments by partly inactivating cofilin. A phosphatase could reactivate cofilin, which would finally accumulate on the filaments, thereby increasing its activity and facilitating the disruption of the F-actin network in the phagocytic cup. (B) Inhibition of endogenous LIMK by expressing the dominant negative LIMK1− or expression of the nonphophorylatable S3A mutant would lead to an excessive activity of cofilin, resulting in increased filament disassembly and abortion of the actin cup formation. (C) Overexpressing LIMK1 would increase cofilin phosphorylation, leading to the inhibition of its actin depolymerizing activity and inducing the formation of a rigid structure beneath the entering particles made of a thick filament network. These events would prevent the engulfment of the particle into the cell, resulting in the inhibition of the phagocytic cup closure.
Similar articles
- WASP-related proteins, Abi1 and Ena/VASP are required for Listeria invasion induced by the Met receptor.
Bierne H, Miki H, Innocenti M, Scita G, Gertler FB, Takenawa T, Cossart P. Bierne H, et al. J Cell Sci. 2005 Apr 1;118(Pt 7):1537-47. doi: 10.1242/jcs.02285. Epub 2005 Mar 15. J Cell Sci. 2005. PMID: 15769844 - InlB-mediated Listeria monocytogenes internalization requires a balanced phospholipase D activity maintained through phospho-cofilin.
Han X, Yu R, Ji L, Zhen D, Tao S, Li S, Sun Y, Huang L, Feng Z, Li X, Han G, Schmidt M, Han L. Han X, et al. Mol Microbiol. 2011 Aug;81(4):860-80. doi: 10.1111/j.1365-2958.2011.07726.x. Epub 2011 Jul 4. Mol Microbiol. 2011. PMID: 21722201 - Efficient Salmonella entry requires activity cycles of host ADF and cofilin.
Dai S, Sarmiere PD, Wiggan O, Bamburg JR, Zhou D. Dai S, et al. Cell Microbiol. 2004 May;6(5):459-71. doi: 10.1111/j.1462-5822.2004.00375.x. Cell Microbiol. 2004. PMID: 15056216 - Lim kinases, regulators of actin dynamics.
Bernard O. Bernard O. Int J Biochem Cell Biol. 2007;39(6):1071-6. doi: 10.1016/j.biocel.2006.11.011. Epub 2006 Nov 28. Int J Biochem Cell Biol. 2007. PMID: 17188549 Review. - Host-pathogen interactions during entry and actin-based movement of Listeria monocytogenes.
Ireton K, Cossart P. Ireton K, et al. Annu Rev Genet. 1997;31:113-38. doi: 10.1146/annurev.genet.31.1.113. Annu Rev Genet. 1997. PMID: 9442892 Review.
Cited by
- Stathmin recruits tubulin to Listeria monocytogenes-induced actin comets and promotes bacterial dissemination.
Costa AC, Carvalho F, Cabanes D, Sousa S. Costa AC, et al. Cell Mol Life Sci. 2019 Mar;76(5):961-975. doi: 10.1007/s00018-018-2977-7. Epub 2018 Dec 1. Cell Mol Life Sci. 2019. PMID: 30506415 Free PMC article. - Epidermal growth factor receptor-PI3K signaling controls cofilin activity to facilitate herpes simplex virus 1 entry into neuronal cells.
Zheng K, Xiang Y, Wang X, Wang Q, Zhong M, Wang S, Wang X, Fan J, Kitazato K, Wang Y. Zheng K, et al. mBio. 2014 Jan 14;5(1):e00958-13. doi: 10.1128/mBio.00958-13. mBio. 2014. PMID: 24425731 Free PMC article. - Actin-interacting and flagellar proteins in Leishmania spp.: Bioinformatics predictions to functional assignments in phagosome formation.
Diniz MC, Costa MP, Pacheco AC, Kamimura MT, Silva SC, Carneiro LD, Sousa AP, Soares CE, Souza CS, de Oliveira DM. Diniz MC, et al. Genet Mol Biol. 2009 Jul;32(3):652-65. doi: 10.1590/S1415-47572009000300033. Epub 2009 Sep 1. Genet Mol Biol. 2009. PMID: 21637533 Free PMC article. - The Chlamydia pneumoniae Tarp Ortholog CPn0572 Stabilizes Host F-Actin by Displacement of Cofilin.
Zrieq R, Braun C, Hegemann JH. Zrieq R, et al. Front Cell Infect Microbiol. 2017 Dec 12;7:511. doi: 10.3389/fcimb.2017.00511. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 29376031 Free PMC article. - In Situ Molecular Ecological Analyses Illuminate Distinct Factors Regulating Formation and Demise of a Harmful Dinoflagellate Bloom.
Yu L, Li T, Li H, Ma M, Li L, Lin S. Yu L, et al. Microbiol Spectr. 2023 Jun 15;11(3):e0515722. doi: 10.1128/spectrum.05157-22. Epub 2023 Apr 19. Microbiol Spectr. 2023. PMID: 37074171 Free PMC article.
References
- Aizawa, H., Y. Fukui, and I. Yahara. 1997. Live dynamics of Dictyostelium cofilin suggests a role in remodeling actin latticework into bundles. J. Cell Sci. 110:2333–2344. - PubMed
- Arber, S., F.A. Barbayannis, H. Hanser, C. Schneider, C.A. Stanyon, O. Bernard, and P. Caroni. 1998. Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature. 393:805–809. - PubMed
- Bamburg, J.R. 1999. Proteins of the ADF/cofilin family: essential regulators of actin dynamics. Annu. Rev. Cell Dev. Biol. 15:185–230. - PubMed
- Bierne, H., S. Dramsi, M.P. Gratacap, C. Randriamampita, G. Carpenter, B. Payrastre, and P. Cossart. 2000. The invasion protein InIB from Listeria monocytogenes activates PLC-γ1 downstream from PI 3-kinase. Cell. Microbiol. 2:465–477. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous