A computational analysis of sequence features involved in recognition of short introns - PubMed (original) (raw)
A computational analysis of sequence features involved in recognition of short introns
L P Lim et al. Proc Natl Acad Sci U S A. 2001.
Abstract
Splicing of short introns by the nuclear pre-mRNA splicing machinery is thought to proceed via an "intron definition" mechanism, in which the 5' and 3' splice sites (5'ss, 3'ss, respectively) are initially recognized and paired across the intron. Here, we describe a computational analysis of sequence features involved in recognition of short introns by using available transcript data from five eukaryotes with complete or nearly complete genomic sequences. The information content of five different transcript features was measured by using methods from information theory, and Monte Carlo simulations were used to determine the amount of information required for accurate recognition of short introns in each organism. We conclude: (i) that short introns in Drosophila melanogaster and Caenorhabditis elegans contain essentially all of the information for their recognition by the splicing machinery, and computer programs that simulate splicing specificity can predict the exact boundaries of approximately 95% of short introns in both organisms; (ii) that in yeast, the 5'ss, branch signal, and 3'ss can accurately identify intron locations but do not precisely determine the location of 3' cleavage in every intron; and (iii) that the 5'ss, branch signal, and 3'ss are not sufficient to accurately identify short introns in plant and human transcripts, but that specific subsets of candidate intronic enhancer motifs can be identified in both human and Arabidopsis that contribute dramatically to the accuracy of splicing simulators.
Figures
Figure 1
Intron length distributions. Histograms of the lengths of introns from each organism are plotted, using a log scale for the abscissa. Each histogram was fitted as a mixture of two lognormal distributions by using the
r
statistical package (curved lines). The position of the point of intersection of these distributions is indicated. S. ce., S. cerevisiae; C. el., C. elegans; D. me., D. melanogaster; A. th.,A. thaliana; H. sa., Homo sapiens.
Figure 2
Splice signal motifs. Sequence motifs for the 5′ss (A), branch site (B), and 3′ss (C) are displayed by using the
pictogram
program (
http://genes.mit.edu/pictogram.html
). The height of each letter is proportional to the frequency of the corresponding base at the given position, and bases are listed in descending order of frequency from top to bottom. The RelEnt (in bits) of the motif model used in our analyses (I1M or WMM) relative to the background transcript base composition is also shown. The splice junctions and branch point are marked by inverted triangles.
Figure 3
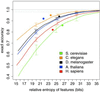
Monte Carlo estimation of information required for short intron recognition. EAc of prediction of short introns by
pairscan
in randomized transcripts is plotted versus the sum of the RelEnts of the splice signal motifs used. Dotted gray line indicates 98% EAc. Each curve is the best-fit from 130 simulations. Brackets indicate 1 SD above and below the best-fit curve for three chosen RelEnt values. Solid circles represent EAc for
intronscan
in real transcripts versus the sum of the RelEnts of the transcript features used.
Figure 4
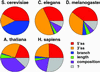
Relative contributions of five transcript features to intron detection. The area of each wedge represents the relative contribution to intron detection accuracy of the corresponding transcript feature, calculated as described in Methods. The sizes of the wedges are scaled so that the complete circle represents the RelEnt per intron required to achieve 98% detection accuracy in each organism, derived from Fig. 3.
Figure 5
Contribution of subsets of pentamers to intron prediction. Exact prediction accuracies are shown for
intronscan
by using the 5′ss and 3′ss signals and specialized intron composition models that score particular subsets of pentamers (see the supporting information) as a function of the number of pentamers used. Circles represent accuracy calculated by using 0, 10, 20, 40, 60, and 100 pentamers, with pentamers chosen in order from high values of_f_log(f/g) to low, where f and g are the pentamer frequency in introns and exons, respectively, using a protocol that avoids choosing overlapping pentamers (see the supporting information). (A) Drosophila, (B)Arabidopsis, (C) human. (D). The first ten intron-biased pentamers chosen from each organism. The dashed black line represents average accuracy for 25 random orderings of pentamers. The solid gray line represents accuracy by using all 1,024 pentamers—dashed gray lines are described in text.
Similar articles
- RNA-Seq approach for accurate characterization of splicing efficiency of yeast introns.
Xia X. Xia X. Methods. 2020 Apr 1;176:25-33. doi: 10.1016/j.ymeth.2019.03.019. Epub 2019 Mar 26. Methods. 2020. PMID: 30926533 - Identification of new branch points and unconventional introns in Saccharomyces cerevisiae.
Gould GM, Paggi JM, Guo Y, Phizicky DV, Zinshteyn B, Wang ET, Gilbert WV, Gifford DK, Burge CB. Gould GM, et al. RNA. 2016 Oct;22(10):1522-34. doi: 10.1261/rna.057216.116. Epub 2016 Jul 29. RNA. 2016. PMID: 27473169 Free PMC article. - RNA Splicing by the Spliceosome.
Wilkinson ME, Charenton C, Nagai K. Wilkinson ME, et al. Annu Rev Biochem. 2020 Jun 20;89:359-388. doi: 10.1146/annurev-biochem-091719-064225. Epub 2019 Dec 3. Annu Rev Biochem. 2020. PMID: 31794245 Review. - Recognition of the 5' splice site by the spliceosome.
Konarska MM. Konarska MM. Acta Biochim Pol. 1998;45(4):869-81. Acta Biochim Pol. 1998. PMID: 10397335 Review.
Cited by
- Mutation of a U2 snRNA gene causes global disruption of alternative splicing and neurodegeneration.
Jia Y, Mu JC, Ackerman SL. Jia Y, et al. Cell. 2012 Jan 20;148(1-2):296-308. doi: 10.1016/j.cell.2011.11.057. Cell. 2012. PMID: 22265417 Free PMC article. - Minimal introns are not "junk".
Yu J, Yang Z, Kibukawa M, Paddock M, Passey DA, Wong GK. Yu J, et al. Genome Res. 2002 Aug;12(8):1185-9. doi: 10.1101/gr.224602. Genome Res. 2002. PMID: 12176926 Free PMC article. - AUG sequences are required to sustain nonsense-codon-mediated suppression of splicing.
Kamhi E, Yahalom G, Kass G, Hacham Y, Sperling R, Sperling J. Kamhi E, et al. Nucleic Acids Res. 2006 Jul 19;34(12):3421-33. doi: 10.1093/nar/gkl390. Print 2006. Nucleic Acids Res. 2006. PMID: 16855285 Free PMC article. - Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation.
Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, Celniker SE, Graveley BR, Lai EC. Westholm JO, et al. Cell Rep. 2014 Dec 11;9(5):1966-1980. doi: 10.1016/j.celrep.2014.10.062. Epub 2014 Nov 26. Cell Rep. 2014. PMID: 25544350 Free PMC article. - The spliceosome: a flexible, reversible macromolecular machine.
Hoskins AA, Moore MJ. Hoskins AA, et al. Trends Biochem Sci. 2012 May;37(5):179-88. doi: 10.1016/j.tibs.2012.02.009. Epub 2012 Apr 3. Trends Biochem Sci. 2012. PMID: 22480731 Free PMC article. Review.
References
- Claverie J M. Genome Res. 2000;10:1277–1279. - PubMed
- International Human Genome Sequencing Consortium. Nature (London) 2001;409:860–921. - PubMed
- Berget S M. J Biol Chem. 1995;270:2411–2414. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases