Induction of cytotoxic T lymphocyte antigen 4 (CTLA-4) restricts clonal expansion of helper T cells - PubMed (original) (raw)
Induction of cytotoxic T lymphocyte antigen 4 (CTLA-4) restricts clonal expansion of helper T cells
A M Doyle et al. J Exp Med. 2001.
Abstract
Cytotoxic T lymphocyte antigen (CTLA)-4 plays an essential role in immunologic homeostasis. How this negative regulator of T cell activation executes its functions has remained controversial. We now provide evidence that CTLA-4 mediates a cell-intrinsic counterbalance to restrict the clonal expansion of proliferating CD4(+) T cells. The regulation of CTLA-4 expression and function ensures that, after approximately 3 cell divisions of expansion, most progeny will succumb to either proliferative arrest or death over the ensuing three cell divisions. The quantitative precision of the counterbalance hinges on the graded, time-independent induction of CTLA-4 expression during the first three cell divisions. In contrast to the limits imposed on unpolarized cells, T helper type 1 (Th1) and Th2 effector progeny may be rescued from proliferative arrest by interleukin (IL)-12 and IL-4 signaling, respectively, allowing appropriately stimulated progeny to proceed to the stage of tissue homing. These results suggest that the cell-autonomous regulation of CTLA-4 induction may be a central checkpoint of clonal expansion of CD4(+) T cells, allowing temporally and spatially restricted growth of progeny to be dictated by the nature of the threat posed to the host.
Figures
Figure 1
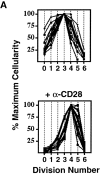
Uniformity in the clonal expansion of CD4+ T cells. (A) CD8-depleted, CFSE-labeled splenocytes were stimulated (see Materials and Methods) with varying concentrations of anti-CD3 mAb (0.05–5.0 μg/ml), and analyzed on day 3, 4, 5 or 6 (top panel). Cellularity of live, CD4+ cells, as determined by specific staining with anti-CD4 mAb, in relation to cell division number is depicted graphically. For comparative purposes the number of cells in each cell division as determined by CFSE dilution (x-axis) was expressed as a percentage of the number of cells in the cell division having the largest number of cells (y-axis) within each experimental condition. Top panel includes 10 experiments (with each line representing one experimental condition and time point). Bottom panel depicts 10 experiments (with each line representing one experimental condition and time point) using varying concentrations of both anti-CD3 (0.01–2.0 μg/ml) and anti-CD28 (0.3–10.0 μg/ml) mAbs, all analyzed on day 4. Cells from both C57BL/6 and BALB/c mice were tested. Dose responses of anti-CD3 and anti-CD28 were tested at least twice. (B) CD8-depleted, CFSE-labeled splenocytes were stimulated with anti-CD3 mAb (1.0 μg/ml), anti-CD28 mAb (2.0 μg/ml), and rIL-2 (2 U/ml) for 3 d, and stained with fluorochrome-conjugated Annexin V and anti-CD4 mAb. Binding of Annexin V (“% Apoptotic,” y-axis) among CD4+ cells of indicated division number (x-axis) is depicted in bar graph (left panel), and flow cytometric data of CD4+ events is displayed (right panel) as cell division (x-axis) versus Annexin V binding (y-axis). Polygonal gate indicates upper limit of background fluorescence. Results are representative of four separate experiments.
Figure 1
Uniformity in the clonal expansion of CD4+ T cells. (A) CD8-depleted, CFSE-labeled splenocytes were stimulated (see Materials and Methods) with varying concentrations of anti-CD3 mAb (0.05–5.0 μg/ml), and analyzed on day 3, 4, 5 or 6 (top panel). Cellularity of live, CD4+ cells, as determined by specific staining with anti-CD4 mAb, in relation to cell division number is depicted graphically. For comparative purposes the number of cells in each cell division as determined by CFSE dilution (x-axis) was expressed as a percentage of the number of cells in the cell division having the largest number of cells (y-axis) within each experimental condition. Top panel includes 10 experiments (with each line representing one experimental condition and time point). Bottom panel depicts 10 experiments (with each line representing one experimental condition and time point) using varying concentrations of both anti-CD3 (0.01–2.0 μg/ml) and anti-CD28 (0.3–10.0 μg/ml) mAbs, all analyzed on day 4. Cells from both C57BL/6 and BALB/c mice were tested. Dose responses of anti-CD3 and anti-CD28 were tested at least twice. (B) CD8-depleted, CFSE-labeled splenocytes were stimulated with anti-CD3 mAb (1.0 μg/ml), anti-CD28 mAb (2.0 μg/ml), and rIL-2 (2 U/ml) for 3 d, and stained with fluorochrome-conjugated Annexin V and anti-CD4 mAb. Binding of Annexin V (“% Apoptotic,” y-axis) among CD4+ cells of indicated division number (x-axis) is depicted in bar graph (left panel), and flow cytometric data of CD4+ events is displayed (right panel) as cell division (x-axis) versus Annexin V binding (y-axis). Polygonal gate indicates upper limit of background fluorescence. Results are representative of four separate experiments.
Figure 2
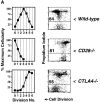
CTLA-4 limits clonal expansion of CD4+ T cells. (A) CFSE-labeled splenocytes from 3–4-wk-old B10.D2 wild-type (top), CD28−/− (middle), and CTLA-4−/− (bottom) DO11.10 TCR transgenic mice were stimulated with anti-CD3 mAb (2.0 μg/ml) for 4 d. For each genotype, computation of relative cellularity of live-gated, CD4+ cells within each cell division was determined as in Fig. 1 A, and is depicted graphically (left column). At the time of harvest, and anti-CD4 staining, cells were also resuspended in propidium iodide for determination of cell death (right column). Flow cytometric data of CFSE dilution (“Cell Division”, x-axis) versus exclusion (lower gate)/inclusion (upper gate) of propidium iodide (y-axis) among all CD4+ events is displayed to the right of cellularity graphs. Numbers above horizontal gates indicate the percentage of dead cells. Results are representative of three separate experiments. (B) CFSE-labeled splenocytes from 5-wk-old littermate BALB/c CTLA-4+/+ (top) and CTLA-4+/− (bottom) DO11.10 TCR transgenic mice were stimulated with anti-CD3 mAb (2.0 μg/ml) for 4 d and analyzed identically to the manner described for part A. Identical results were obtained with cells from B10.D2 littermate-matched pairs of CTLA-4+/+ and CTLA-4+/− mice. Results are representative of four separate experiments.
Figure 2
CTLA-4 limits clonal expansion of CD4+ T cells. (A) CFSE-labeled splenocytes from 3–4-wk-old B10.D2 wild-type (top), CD28−/− (middle), and CTLA-4−/− (bottom) DO11.10 TCR transgenic mice were stimulated with anti-CD3 mAb (2.0 μg/ml) for 4 d. For each genotype, computation of relative cellularity of live-gated, CD4+ cells within each cell division was determined as in Fig. 1 A, and is depicted graphically (left column). At the time of harvest, and anti-CD4 staining, cells were also resuspended in propidium iodide for determination of cell death (right column). Flow cytometric data of CFSE dilution (“Cell Division”, x-axis) versus exclusion (lower gate)/inclusion (upper gate) of propidium iodide (y-axis) among all CD4+ events is displayed to the right of cellularity graphs. Numbers above horizontal gates indicate the percentage of dead cells. Results are representative of three separate experiments. (B) CFSE-labeled splenocytes from 5-wk-old littermate BALB/c CTLA-4+/+ (top) and CTLA-4+/− (bottom) DO11.10 TCR transgenic mice were stimulated with anti-CD3 mAb (2.0 μg/ml) for 4 d and analyzed identically to the manner described for part A. Identical results were obtained with cells from B10.D2 littermate-matched pairs of CTLA-4+/+ and CTLA-4+/− mice. Results are representative of four separate experiments.
Figure 3
Survival and proliferation are antagonized in clonally expanding CD4+ T cells. 4 d after stimulation, cells were stained with anti-CD4 mAb and resuspended in propidium iodide for determination of cell death. Flow cytometric data of CFSE dilution (“Cell Division”, x-axis) versus exclusion/inclusion of propidium iodide (y-axis) among all CD4+ events are displayed. Numbers above horizontal gates indicate the percentage of dead cells as determined by uptake of propidium iodide. (A) CFSE-labeled splenocytes from 6-wk-old BALB/c (from left to right) wild-type, Bcl-xL-transgenic, and CD28−/− DO11.10 TCR transgenic mice were stimulated with anti-CD3 mAb (2.0 μg/ml) for 4 d. Where indicated (far right panel), human rIL-2 (50 U/ml) was added. (B) In a separate experiment, splenocytes from 6-wk-old C57BL/6 mice were either CD8-depleted (left) or subjected to positive selection (>98% purity) of CD4+ cells (right) before labeling with CFSE and stimulation with either soluble anti-CD3 mAb and anti-CD28 mAb (2.0 μg/ml) plus human rIL-2 (5 U/ml) (left) or plate-bound anti-CD3 mAb and soluble anti-CD28 mAb (2.0 μg/ml) plus human rIL-2 (5 U/ml) (right, “no APCs”). All results are representative of at least two separate experiments.
Figure 4
Cell cycle–coupled induction of CTLA-4. (A–E) C57BL/6, CD8-depleted, CFSE-labeled splenocytes were stimulated with anti-CD3 (2.0 μg/ml) for 3 d. All flow cytometric plots depict only live-gated, CD4+ events. Polygonal gates are drawn around the upper limit of background staining, as illustrated in part A. (A) After culture in the absence (left) or presence (right) of mimosine (300 μM), cells were fixed, permeabilized, and stained (see Materials and Methods) with anti-CD4 mAb and either fluorochrome-conjugated hamster control mAb (top row) or hamster anti–mouse CTLA-4 mAb (lower row), before analysis of cell division (x-axis) versus control staining (top) or total cellular CTLA-4 expression (bottom) (y-axis). (B) Cells were washed and stained (without prior fixation or permeabilization) with either anti-CD25, anti-CD44, or control mAb, before analysis of cell division (x-axis) versus surface expression (y-axis) of CD25 (left panels) and CD44 (right panels). (C) Cells were washed, fixed and either stained directly (“surface,” left panel) or permeabilized before staining (“total,” right panel) as above. (D) In the same experiment as C, a group of cells was stimulated in the presence of sodium butyrate (600 μM), to inhibit histone deacetylases, and analyzed for total cellular CTLA-4 expression (y-axis). (E) Cytochalasin B (3.5 μg/ml) was used to prevent cytokinesis in stimulated cells (left panel) before analysis of both DNA content (x-axis, right panel) and CTLA-4 expression (y-axis). (F) Splenocytes from 4 week-old BALB/c CTLA-4+/+ (filled symbols) and CTLA-4+/− (open symbols) littermate mice were stimulated and analyzed as in part A. Geometric mean fluorescence intensity of CTLA-4 expression (y-axis) for each cell generation (x-axis) is displayed. All experiments were performed at least twice.
Figure 5
Growth signals can counteract intrinsic limits on clonal expansion. In all experiments, T cells were stimulated for 4 d. All flow cytometric plots depict only live-gated, CD4+ events. Vertical gates are drawn before the fifth cell division, and percentage of cells that achieved five or more divisions is indicated in left top or bottom corner. (A) CFSE-labeled splenocytes from 3–4-wk-old B10.D2 CTLA4+/+ (top panel) and CTLA-4−/− (bottom panel) DO11.10 TCR transgenic mice were stimulated with anti-CD3 (2.0 μg/ml), before staining with anti-CD4 mAb, and analysis of CFSE dilution (“Cell Division,” x-axis) versus forward light scatter (“Cell Size,” y-axis). (B) CD8-depleted, CFSE-labeled splenocytes were stimulated with anti-CD3 in the indicated doses of human rIL-2 before analysis as in part A. (C) CD8-depleted, CFSE-labeled splenocytes were stimulated with anti-CD3 (2.0 μg/ml), anti-CD28 (0.5 μg/ml), rIL-2 (10 U/ml), and either rIL-12 (5 ng/ml) plus anti–IL-4 (10 μg/ml) (“Th1”conditions, left) or rIL-4 (5 U/ml) plus anti–IL-12 (10 μg/ml) (“Th2” conditions, right) before analysis of intracellular cytokine staining as described in Materials and Methods. Cell division (x-axis) and intracellular accumulation (y-axis) of IFN-γ (left) or IL-4 (right) are depicted in the top row inset. Horizontal gates indicate level of background staining. Cells that were unambiguously cytokine-negative (middle panels) or cytokine-positive (bottom panels) were further analyzed for cell division (x-axis) and cell size (y-axis) as in part A. (D) Expression of CCR7 (top panels) and HPRT (bottom panels) was determined by RT-PCR among indicated cell division numbers (“Div. No.”), as described in Materials and Methods. All results are representative of at least three separate experiments.
Figure 6
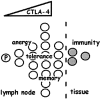
CTLA-4 as the central checkpoint of clonal expansion. Speculative model in which limits imposed on clonal expansion of a precursor (“P”) CD4+ T cell by CTLA-4 expression and function can mediate various immunologic outcomes. Progressive induction of CTLA-4 during the initial cell divisions (triangle of dose response) would maintain homeostasis (manifest as anergy or tolerance) in the absence of inflammatory signals by limiting the division of clonal progeny. In response to a pathogen, a subset of clonal progeny may receive growth and maturation signals that cause further division, enabling their emigration from lymph nodes. Pathogen-specific clonal progeny that arrest before the fifth division could remain in lymphoid niches that may support long-term survival (memory), perhaps to be rapidly mobilized into further division and emigration during reinfection.
Similar articles
- Engagement of cytotoxic T lymphocyte-associated antigen 4 (CTLA-4) induces transforming growth factor beta (TGF-beta) production by murine CD4(+) T cells.
Chen W, Jin W, Wahl SM. Chen W, et al. J Exp Med. 1998 Nov 16;188(10):1849-57. doi: 10.1084/jem.188.10.1849. J Exp Med. 1998. PMID: 9815262 Free PMC article. - Blockade of CTLA-4 signals inhibits Th2-mediated murine chronic graft-versus-host disease by an enhanced expansion of regulatory CD8+ T cells.
Sakurai J, Ohata J, Saito K, Miyajima H, Hirano T, Kohsaka T, Enomoto S, Okumura K, Azuma M. Sakurai J, et al. J Immunol. 2000 Jan 15;164(2):664-9. doi: 10.4049/jimmunol.164.2.664. J Immunol. 2000. PMID: 10623808 - Polarization of naive CD4+ T cells toward the Th1 subset by CTLA-4 costimulation.
Kato T, Nariuchi H. Kato T, et al. J Immunol. 2000 Apr 1;164(7):3554-62. doi: 10.4049/jimmunol.164.7.3554. J Immunol. 2000. PMID: 10725710 - CTLA-4, a negative regulator of T-lymphocyte activation.
Waterhouse P, Marengère LE, Mittrücker HW, Mak TW. Waterhouse P, et al. Immunol Rev. 1996 Oct;153:183-207. doi: 10.1111/j.1600-065x.1996.tb00925.x. Immunol Rev. 1996. PMID: 9010724 Review. No abstract available. - The role of CTLA-4 in the regulation and initiation of T-cell responses.
Chambers CA, Krummel MF, Boitel B, Hurwitz A, Sullivan TJ, Fournier S, Cassell D, Brunner M, Allison JP. Chambers CA, et al. Immunol Rev. 1996 Oct;153:27-46. doi: 10.1111/j.1600-065x.1996.tb00919.x. Immunol Rev. 1996. PMID: 9010718 Review. No abstract available.
Cited by
- Signaling pathways and targeted therapies in lung squamous cell carcinoma: mechanisms and clinical trials.
Niu Z, Jin R, Zhang Y, Li H. Niu Z, et al. Signal Transduct Target Ther. 2022 Oct 5;7(1):353. doi: 10.1038/s41392-022-01200-x. Signal Transduct Target Ther. 2022. PMID: 36198685 Free PMC article. Review. - Multiple functions for CD28 and cytotoxic T lymphocyte antigen-4 during different phases of T cell responses: implications for arthritis and autoimmune diseases.
Brunner-Weinzierl MC, Hoff H, Burmester GR. Brunner-Weinzierl MC, et al. Arthritis Res Ther. 2004;6(2):45-54. doi: 10.1186/ar1158. Epub 2004 Mar 3. Arthritis Res Ther. 2004. PMID: 15059264 Free PMC article. Review. - Infection of CD127+ (interleukin-7 receptor+) CD4+ cells and overexpression of CTLA-4 are linked to loss of antigen-specific CD4 T cells during primary human immunodeficiency virus type 1 infection.
Zaunders JJ, Ip S, Munier ML, Kaufmann DE, Suzuki K, Brereton C, Sasson SC, Seddiki N, Koelsch K, Landay A, Grey P, Finlayson R, Kaldor J, Rosenberg ES, Walker BD, Fazekas de St Groth B, Cooper DA, Kelleher AD. Zaunders JJ, et al. J Virol. 2006 Oct;80(20):10162-72. doi: 10.1128/JVI.00249-06. J Virol. 2006. PMID: 17005693 Free PMC article. - Immune checkpoints, their control by immunotherapy and ovarian cancer.
Bose CK. Bose CK. Contemp Oncol (Pozn). 2017;21(3):189-196. doi: 10.5114/wo.2017.70108. Epub 2017 Sep 29. Contemp Oncol (Pozn). 2017. PMID: 29180924 Free PMC article. Review. - Checkpoint Inhibitors for Non-Small Cell Lung Cancer Among Older Adults.
Elias R, Morales J, Presley C. Elias R, et al. Curr Oncol Rep. 2017 Sep;19(9):62. doi: 10.1007/s11912-017-0619-0. Curr Oncol Rep. 2017. PMID: 28755314 Review.
References
- Janeway C.A., Bottomly K. Signals and signs for lymphocyte responses. Cell. 1994;76:275–285. - PubMed
- Lanzavecchia A., Sallusto F. Dynamics of T lymphocyte responsesintermediates, effectors, and memory cells. Science. 2000;290:92–97. - PubMed
- Reiner S.L., Seder R.A. Dealing from the evolutionary pawnshophow lymphocytes make decisions. Immunity. 1999;11:1–10. - PubMed
- Gett A.V., Hodgkin P.D. A cellular calculus for signal integration by T cells. Nat. Immunol. 2000;1:239–244. - PubMed
- Ucker D.S., Ashwell J.D., Nickas G. Activation-driven T cell death. I. Requirements for de novo transcription and translation and association with genome fragmentation. J. Immunol. 1989;143:3461–3469. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials