Physical and functional interactions of human DNA polymerase eta with PCNA - PubMed (original) (raw)
Physical and functional interactions of human DNA polymerase eta with PCNA
L Haracska et al. Mol Cell Biol. 2001 Nov.
Abstract
Human DNA polymerase eta (hPoleta) functions in the error-free replication of UV-damaged DNA, and mutations in hPoleta cause cancer-prone syndrome, the variant form of xeroderma pigmentosum. However, in spite of its key role in promoting replication through a variety of distorting DNA lesions, the manner by which hPoleta is targeted to the replication machinery stalled at a lesion site remains unknown. Here, we provide evidence for the physical interaction of hPoleta with proliferating cell nuclear antigen (PCNA) and show that mutations in the PCNA binding motif of hPoleta inactivate this interaction. PCNA, together with replication factor C and replication protein A, stimulates the DNA synthetic activity of hPoleta, and steady-state kinetic studies indicate that this stimulation accrues from an increase in the efficiency of nucleotide insertion resulting from a reduction in the apparent K(m) for the incoming nucleotide.
Figures
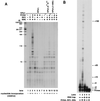
FIG. 1
PCNA binding motif of hPolη. (A) C-terminal amino acids 699 to 712 of hPolη are aligned with the PCNA binding motifs identified in various PCNA binding proteins. The highly conserved residues are shown in bold. Hs, human; Sc, S. cerevisiae. (B) Mutations made in the PCNA binding motif of hPolη. In the schematic representation of hPolη, the five highly conserved motifs (I to V) shared among different members of the Polη/UmuC/DinB protein family are indicated, and the C2H2 motif conserved in the Polη family is shown (20). The amino acid residues present in the extreme C-terminal region are shown; the amino acids highly conserved in the consensus PCNA binding motif are shown in bold. In the hPolη (1-695) mutant protein, the last 18 amino acids from the C terminus were deleted (indicated by an arrow). In the hPolη A707-A708 mutant protein, the F residues at positions 707 and 708 (indicated by asterisks) were changed to A residues.
FIG. 2
Polη forms a complex with PCNA. Six-His–hPCNA (4 μg) was mixed either with wild-type GST-hPolη protein (lanes 1 to 3 and 13 to 15) or with mutant GST-hPolη A707-A708 (lanes 4 to 6 and 16 to 18) or GST-hPolη (1-695) (lanes 7 to 9 and 19 to 21) protein (4 μg each). As controls, the same amounts of GST-hPolη (lanes 10 to 12) or six-His–hPCNA (lanes 22 to 24) protein were used alone. After incubation, samples were bound to Ni-NTA (lanes 1 to 12) or glutathione-Sepharose (Seph.) (lanes 13 to 24) beads, followed by washing and elution of the bound proteins by imidazole- or glutathione-containing buffer, respectively. Aliquots of each sample before loading on the beads (L), the flowthrough and wash (F), and the eluted proteins (E) were precipitated by TCA and analyzed on a sodium dodecyl sulfate–12% polyacrylamide gel stained with Coomassie blue. The positions of GST-hPolη and six-His–hPCNA are indicated on the right.
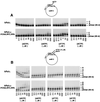
FIG. 3
Stimulation of DNA synthetic activity of hPolη by PCNA. (A) DNA synthesis by the wild-type or PCNA binding site mutant hPolη proteins in the presence or absence of PCNA, RFC, and RPA. The reaction mixtures contained either the wild type hPolη protein (lanes 1 to 8) or the mutant hPolη A707-A708 (lanes 9 and 10) or hPolη (1-695) (lanes 11 and 12) proteins (10 ng each) along with singly primed M13 single-stranded DNA (25 ng), all four dNTPs (100 μM each), [α-32P]dATP, PCNA (100 ng), RFC (50 ng), or RPA (250 ng) or combinations of these proteins. No hPolη was added in lane 13. The amount of DNA synthesis is indicated at the bottom as the relative nucleotide (nt) incorporation. _Hae_III-digested ϕX174 DNA labeled with polynucleotide kinase is shown on the left as a molecular size marker. (B) Processivity of hPolη in the presence of PCNA, RFC, and RPA. hPolη (10 ng) alone (lanes 1 and 4) or in the presence of PCNA (100 ng), RFC (50 ng), and RPA (250 ng) (lanes 2 and 3) was preincubated with a circular single-stranded M13 template DNA (50 ng) singly primed with a 5′ 32P-labeled oligonucleotide for 5 min at 37°C. Primer extension reactions were initiated by adding all four dNTPs (500 μM each) (lanes 1 and 2) or all four dNTPs and excess sonicated herring sperm DNA (0.5 mg/ml) as a trap (lanes 3 and 4). After incubation for10 min at 37°C , samples were quenched and run on a 10% polyacrylamide gel. To demonstrate the effectiveness of the trap, hPolη, along with PCNA, RFC, and RPA, was preincubated with the trap DNA and the primer-template substrate before the addition of dNTPs (lane 5).
FIG. 4
PCNA stimulates deoxynucleotide incorporation by hPolη. (A) Steady-state kinetics of deoxynucleotide incorporation opposite an A residue by hPolη in the presence or absence of PCNA, RFC, and RPA in a standing-start reaction. A portion of the DNA substrate is shown at the top. Polη (10 ng) was incubated with a singly primed circular single-stranded M13 (ssM13) DNA substrate (25 ng) and increasing concentrations of a single deoxynucleotide in the absence or presence of PCNA (100 ng), RFC (50 ng), and RPA (250 ng). The nucleotide (nt) incorporation rate was plotted against the dNTP concentration, and the data were fit to the Michaelis-Menten equation describing a hyperbola. The apparent K m and _V_max values were obtained from the fit and used to calculate the efficiency of deoxynucleotide incorporation (_V_max/K m). (B) Steady-state kinetics of deoxynucleotide incorporation opposite a template C residue by hPolη in the presence or absence of PCNA, RFC, and RPA in a running-start reaction. Reactions were carried out as described for panel A, except that each reaction also included the addition of dTTP (15 μM).
Similar articles
- Interaction with PCNA is essential for yeast DNA polymerase eta function.
Haracska L, Kondratick CM, Unk I, Prakash S, Prakash L. Haracska L, et al. Mol Cell. 2001 Aug;8(2):407-15. doi: 10.1016/s1097-2765(01)00319-7. Mol Cell. 2001. PMID: 11545742 - Proliferating cell nuclear antigen-dependent coordination of the biological functions of human DNA polymerase iota.
Vidal AE, Kannouche P, Podust VN, Yang W, Lehmann AR, Woodgate R. Vidal AE, et al. J Biol Chem. 2004 Nov 12;279(46):48360-8. doi: 10.1074/jbc.M406511200. Epub 2004 Sep 1. J Biol Chem. 2004. PMID: 15342632 - Targeting of human DNA polymerase iota to the replication machinery via interaction with PCNA.
Haracska L, Johnson RE, Unk I, Phillips BB, Hurwitz J, Prakash L, Prakash S. Haracska L, et al. Proc Natl Acad Sci U S A. 2001 Dec 4;98(25):14256-61. doi: 10.1073/pnas.261560798. Epub 2001 Nov 27. Proc Natl Acad Sci U S A. 2001. PMID: 11724965 Free PMC article. - Regulatory role of ubiquitin in eukaryotic DNA translesion synthesis.
Yang K, Weinacht CP, Zhuang Z. Yang K, et al. Biochemistry. 2013 May 14;52(19):3217-28. doi: 10.1021/bi400194r. Epub 2013 May 1. Biochemistry. 2013. PMID: 23634825 Review. - Molecular mechanism of PCNA-dependent base excision repair.
Matsumoto Y. Matsumoto Y. Prog Nucleic Acid Res Mol Biol. 2001;68:129-38. doi: 10.1016/s0079-6603(01)68095-4. Prog Nucleic Acid Res Mol Biol. 2001. PMID: 11554292 Review.
Cited by
- Different types of interaction between PCNA and PIP boxes contribute to distinct cellular functions of Y-family DNA polymerases.
Masuda Y, Kanao R, Kaji K, Ohmori H, Hanaoka F, Masutani C. Masuda Y, et al. Nucleic Acids Res. 2015 Sep 18;43(16):7898-910. doi: 10.1093/nar/gkv712. Epub 2015 Jul 13. Nucleic Acids Res. 2015. PMID: 26170230 Free PMC article. - Controlling the subcellular localization of DNA polymerases iota and eta via interactions with ubiquitin.
Plosky BS, Vidal AE, Fernández de Henestrosa AR, McLenigan MP, McDonald JP, Mead S, Woodgate R. Plosky BS, et al. EMBO J. 2006 Jun 21;25(12):2847-55. doi: 10.1038/sj.emboj.7601178. Epub 2006 Jun 8. EMBO J. 2006. PMID: 16763556 Free PMC article. - Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination.
Watanabe K, Tateishi S, Kawasuji M, Tsurimoto T, Inoue H, Yamaizumi M. Watanabe K, et al. EMBO J. 2004 Oct 1;23(19):3886-96. doi: 10.1038/sj.emboj.7600383. Epub 2004 Sep 9. EMBO J. 2004. PMID: 15359278 Free PMC article. - Proliferating cell nuclear antigen inhibitors block distinct stages of herpes simplex virus infection.
Packard JE, Williams MR, Fromuth DP, Dembowski JA. Packard JE, et al. PLoS Pathog. 2023 Jul 24;19(7):e1011539. doi: 10.1371/journal.ppat.1011539. eCollection 2023 Jul. PLoS Pathog. 2023. PMID: 37486931 Free PMC article. - FF483-484 motif of human Polη mediates its interaction with the POLD2 subunit of Polδ and contributes to DNA damage tolerance.
Baldeck N, Janel-Bintz R, Wagner J, Tissier A, Fuchs RP, Burkovics P, Haracska L, Despras E, Bichara M, Chatton B, Cordonnier AM. Baldeck N, et al. Nucleic Acids Res. 2015 Feb 27;43(4):2116-25. doi: 10.1093/nar/gkv076. Epub 2015 Feb 6. Nucleic Acids Res. 2015. PMID: 25662213 Free PMC article.
References
- Bailly V, Lauder S, Prakash S, Prakash L. Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. J Biol Chem. 1997;272:23360–23365. - PubMed
- Bambara R A, Murante R S, Henricksen L A. Enzymes and reactions at the eukaryotic DNA replication fork. J Biol Chem. 1997;272:4647–4650. - PubMed
- Bloom L B, Chen X, Fygenson D K, Turner J, O'Donnell M, Goodman M F. Fidelity of Escherichia coli DNA polymerase III holoenzyme The effects of β, γ complex processivity proteins and ɛ proofreading exonuclease on nucleotide misincorporation efficiencies. J Biol Chem. 1997;272:27919–27930. - PubMed
- Brash D E. Sunlight and the onset of skin cancer. Trends Genet. 1997;13:410–414. - PubMed
- Cai J, Gibbs E, Uhlmann F, Phillips B, Yano N, O'Donnell M, Hurwitz J. A complex consisting of human replication factor C, p40, p37, and p36 subunits is a DNA-dependent ATPase and an intermediate in the assembly of the holoenzyme. J Biol Chem. 1997;272:18974–18981. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous