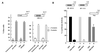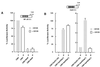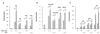Raf kinase inhibitor protein interacts with NF-kappaB-inducing kinase and TAK1 and inhibits NF-kappaB activation - PubMed (original) (raw)
Raf kinase inhibitor protein interacts with NF-kappaB-inducing kinase and TAK1 and inhibits NF-kappaB activation
K C Yeung et al. Mol Cell Biol. 2001 Nov.
Abstract
The Raf kinase inhibitor protein (RKIP) acts as a negative regulator of the mitogen-activated protein (MAP) kinase (MAPK) cascade initiated by Raf-1. RKIP inhibits the phosphorylation of MAP/extracellular signal-regulated kinase 1 (MEK1) by Raf-1 by disrupting the interaction between these two kinases. We show here that RKIP also antagonizes the signal transduction pathways that mediate the activation of the transcription factor nuclear factor kappa B (NF-kappaB) in response to stimulation with tumor necrosis factor alpha (TNF-alpha) or interleukin 1 beta. Modulation of RKIP expression levels affected NF-kappaB signaling independent of the MAPK pathway. Genetic epistasis analysis involving the ectopic expression of kinases acting in the NF-kappaB pathway indicated that RKIP acts upstream of the kinase complex that mediates the phosphorylation and inactivation of the inhibitor of NF-kappaB (IkappaB). In vitro kinase assays showed that RKIP antagonizes the activation of the IkappaB kinase (IKK) activity elicited by TNF-alpha. RKIP physically interacted with four kinases of the NF-kappaB activation pathway, NF-kappaB-inducing kinase, transforming growth factor beta-activated kinase 1, IKKalpha, and IKKbeta. This mode of action bears striking similarities to the interactions of RKIP with Raf-1 and MEK1 in the MAPK pathway. Emerging data from diverse organisms suggest that RKIP and RKIP-related proteins represent a new and evolutionarily highly conserved family of protein kinase regulators. Since the MAPK and NF-kappaB pathways have physiologically distinct roles, the function of RKIP may be, in part, to coordinate the regulation of these pathways.
Figures
FIG. 1
Ablation of RKIP activates and overexpression of RKIP represses basal NF-κB activity. (A) Ablation of RKIP activity by antibody injection activates an NF-κB-dependent reporter (E-sel) but not a cyclic AMP-stimulated reporter (CRE × 5). E-sel is an E-selectin promoter DNA fragment containing one copy of the consensus NF-κB binding site (71). Quiescent Rat1 cells were microinjected with the indicated reporter plasmids and antibodies and either left unstimulated or treated with 20 μg of forskolin per ml, an activator of adenyl cyclase. (B) Overexpression of RKIP represses NF-κB- and AP-1-dependent reporters. RKIP was placed under the control of the cytomegalovirus (CMV) promoter (CMV-RKIP). RKIP or an empty vector control (CMV) was cotransfected with the indicated reporter plasmids into exponentially growing NIH 3T3 cells, and 48 h later extracts were assayed for luciferase activity. The activities of reporters in combination with the empty CMV vector were set to 100%. In all panels the means and standard deviations of at least two independent experiments are shown.
FIG. 2
RKIP can inhibit NF-κB independently of MEK. (A) Pharmacological inhibition of MEK activity does not interfere with the ability of RKIP to repress basal NF-κB activity. 293 cells were cotransfected with an empty vector (cytomegalovirus [CMV]), an RKIP expression vector (CMV-RKIP), and a NF-κB reporter (NF-κB × 3Luc) as indicated. Twenty-four hours after transfection, the medium was replaced with medium containing 10 μM MEK inhibitor U0126 (lanes 2 and 4) or an equivalent volume of carrier (dimethyl sulfoxide) (lanes 1 and 3). Cells were harvested 24 h later, and luciferase activity was determined. Repression was calculated relative to the activity elicited by the reporter plus empty CMV vector (lane 1), which was assigned a value of 100%. (B) Control experiment performed in parallel to demonstrate that U0126 was active under the conditions used. A Gal4 (DNA-binding domain)-Sap1 (transactivation domain) fusion protein [CMV-Gal4(Sap1)] was used in conjunction with a Gal4 (DNA-binding site) reporter (Gal4 × 4 Luc). The Sap1 transactivation domain is a known target of ERK. Activation (lane 5) was measured relative to a Gal4-only vector [CMV-Gal4(1 to 94)] (lane 4) and was set to 1. To control for nonspecific effects of U0126, a Gal4-Sp1 (transactivation domain) fusion protein [CMV-Gal4(Sp1)] was used. This transactivation domain is known to be independent of ERK activity and was not inhibited by U0126 (lanes 1 to 3).
FIG. 3
Coimmunoprecipitation of NIK, TAK1, IKKα, and IKKβ with RKIP. (A) FLAG-RKIP was cotransfected into COS-1 cells with the indicated HA-tagged expression plasmids. Extracts were immunoprecipitated (IP) with anti-HA or anti-FLAG antibodies as indicated. Immunoprecipitates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to membranes, and the expression levels of HA-tagged or FLAG-tagged proteins were monitored by immunoblotting (IB) with the indicated antibodies. (B) HA-RKIP was cotransfected into COS-1 cells with the indicated FLAG-tagged expression plasmids. Extracts were analyzed as indicated above. (C) HA-RKIP was cotransfected into COS-1 cells either alone or with FLAG-tagged IKKα or IKKβ expression plasmids. Extracts were analyzed as indicated above.
FIG. 4
RKIP inhibits NIK- and TAK1-mediated activation of NF-κB. 293 cells were cotransfected with an NF-κB luciferase reporter (NF-κB × 3), an RKIP expression vector (CMV-RKIP), or an empty vector control (cytomegalovirus [CMV]) and expression vectors for NIK, NAK, TAK1, MEKK1, IKKα, IKKβ, and NF-κB/p65 as indicated. The NIK and NAK (A), IKKα, IKKβ, and NF-κB/p65 (B), and TAK1 and MEKK1 (C) vectors were each transfected at two DNA concentrations: 0.1 and 0.5 μg per 35-mm-diameter plate. The NF-κB × 3, CMV-RKIP, and CMV vectors were held constant (50 ng for NF-κB × 3 and 2 μg for CMV-RKIP and CMV). The total DNA concentration per plate was kept constant with pUC19 plasmid DNA. Activity elicited by NF-κB × 3 cotransfected with the CMV vector control in the absence of activating kinases was set to 1. At 2 days posttransfection, cell extracts were prepared and analyzed for luciferase activity. All activities were normalized on the basis of an internal transfection control (thymidine kinase promoter-driven Renilla luciferase reporter).
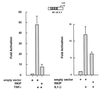
FIG. 5
RKIP inhibits TNF-α- and IL-1β-mediated activation of NF-κB. 293 (A) or 293/IL-1R1 (B) cells were transfected with an NF-κB luciferase reporter (NF-κB × 3) and an RKIP expression vector (CMV-RKIP) or empty vector control (cytomegalovirus [CMV]). Thirty hours after transfection, the cells were either left untreated or were stimulated for 6 h with TNF-α (A) or IL-1β (B) and extracts were prepared and analyzed for luciferase activity. The data presented in the figure are representative of the three experiments we performed.
FIG. 6
Effects of RKIP on the phosphorylation of IκB proteins by IKKα and IKKβ. (A) Inhibition of IκBα phosphorylation by IKKα and IKKβ. 293 cells were transfected with the indicated FLAG-tagged IKK expression vectors with or without an RKIP expression vector. Thirty hours after transfection, cells were either left untreated or were stimulated for 10 min with TNF-α. IKK proteins were immunoprecipitated with anti-FLAG antibody and assayed for kinase activity using purified recombinant bacterially expressed full-length IκBα and [γ-32P]ATP as substrates. (B) Inhibition of IKKα and IKKβ autophosphorylation. 293 cells were transfected as described for panel A. The autophosphorylation of IKK was assayed by incubating the IKKα or IKKβ immunoprecipitates with [γ-32P]ATP. The amount of total IKK protein in each assay was determined by immunoblotting the kinase reactions with polyclonal anti-FLAG antibody. (C) Inhibition of IKK activity by RKIP in vitro. COS-1 cells were transiently cotransfected with FLAG-IKKα (1.5 μg), FLAG-IKKα K44A (1.5 μg), FLAG-IKKβ (2 μg), or FLAG-IKKβ K44A (2 μg), serum deprived for 24 h, and subsequently treated with 50 ng of TNF-α/ml for 10 min. FLAG-tagged proteins were immunoprecipitated with FLAG-M2 antibody, and aliquots of the immunoprecipitate (IP) (10 μl) were incubated on ice for 30 min with increasing concentrations of RKIP (2.5 to 22.5 μM) or 22.5 μM bovine serum albumin. The kinase reaction was initiated by adding 10 μl of a mix containing 2.5 μM GST-IκB (amino acids to 55), 50 μM ATP, and 3 μCi of [32P]ATP in 1× kinase buffer. IKKα reactions were incubated for 10 min at 30°C, and IKKβ reactions were incubated for 30 min. (D) RKIP interferes with TNF-α-induced degradation of IκBα in vivo. 293 cells were transfected with the indicated expression vectors. Thirty hours after transfection, cells were either left untreated or were stimulated for 10 min with TNF-α. Cell lysates were immunoblotted with either anti-FLAG or anti-RKIP antibodies.
FIG. 7
Coimmunoprecipitation of endogenous TAK1 and NIK with RKIP. (A) Coimmunoprecipitation of RKIP and TAK1 from extracts of Rat1 fibroblasts. Rat1 cells (2 × 107) were lysed by sonication in phosphate-buffered saline. Protein extracts were immunoprecipitated with either anti-RKIP or control antibodies. Immunoprecipitated proteins were monitored by immunoblotting (IB) with either anti-RKIP or anti-TAK1 antibodies. The band present in the lane of the RKIP lysate is not IgG but comes from a protein that is cross-reactive with the RKIP antibody. (B) Association of TAK1 with RKIP after IL-1β treatment. 293/IL-1R1 cells were transfected with an HA-RKIP expression vector. Thirty hours after transfection, cells were serum deprived for 24 h and treated with IL-1β as indicated. Cells were harvested in hypotonic buffer and were lysed by rapid expulsion through a 25-gauge hypodermic needle. HA antibody-immunoprecipitated proteins were monitored by immunoblotting with either anti-RKIP or anti-TAK1 antibodies. (C) Fractionation of S100 cytosolic extracts. S100 extracts prepared from Rat1 cells stably transfected with HA-tagged RKIP were fractionated by DEAE Sepharose chromatography as described in Materials and Methods. Fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by immunoblotting with the indicated antibodies. FT, column flowthrough. (D) Coimmunoprecipitation of RKIP, NIK, and TAK1 from DEAE Sepharose chromatography fractions. Fractions shown to contain NIK, TAK1, and HA-RKIP by immunoblotting (shown in panel C) were pooled and immunoprecipitated with either anti-HA or control anti-FLAG antibodies. The precipitated proteins were monitored by immunoblotting with the indicated antibodies.
Similar articles
- Suppression of Raf-1 kinase activity and MAP kinase signalling by RKIP.
Yeung K, Seitz T, Li S, Janosch P, McFerran B, Kaiser C, Fee F, Katsanakis KD, Rose DW, Mischak H, Sedivy JM, Kolch W. Yeung K, et al. Nature. 1999 Sep 9;401(6749):173-7. doi: 10.1038/43686. Nature. 1999. PMID: 10490027 - Raf-1 kinase inhibitor protein: structure, function, regulation of cell signaling, and pivotal role in apoptosis.
Odabaei G, Chatterjee D, Jazirehi AR, Goodglick L, Yeung K, Bonavida B. Odabaei G, et al. Adv Cancer Res. 2004;91:169-200. doi: 10.1016/S0065-230X(04)91005-6. Adv Cancer Res. 2004. PMID: 15327891 Review. - Protein kinase C switches the Raf kinase inhibitor from Raf-1 to GRK-2.
Lorenz K, Lohse MJ, Quitterer U. Lorenz K, et al. Nature. 2003 Dec 4;426(6966):574-9. doi: 10.1038/nature02158. Nature. 2003. PMID: 14654844 - Raf kinase inhibitory protein regulates aurora B kinase and the spindle checkpoint.
Eves EM, Shapiro P, Naik K, Klein UR, Trakul N, Rosner MR. Eves EM, et al. Mol Cell. 2006 Aug;23(4):561-74. doi: 10.1016/j.molcel.2006.07.015. Mol Cell. 2006. PMID: 16916643 Free PMC article. - Regulation and function of IKK and IKK-related kinases.
Häcker H, Karin M. Häcker H, et al. Sci STKE. 2006 Oct 17;2006(357):re13. doi: 10.1126/stke.3572006re13. Sci STKE. 2006. PMID: 17047224 Review.
Cited by
- Cross-Talking Pathways of Rapidly Accelerated Fibrosarcoma-1 (RAF-1) in Alzheimer's Disease.
Hong H, Yu L, Cong W, Kang K, Gao Y, Guan Q, Meng X, Zhang H, Zhou Z. Hong H, et al. Mol Neurobiol. 2024 May;61(5):2798-2807. doi: 10.1007/s12035-023-03765-2. Epub 2023 Nov 8. Mol Neurobiol. 2024. PMID: 37940778 - The inhibitor effect of RKIP on inflammasome activation and inflammasome-dependent diseases.
Qin Q, Liu H, Shou J, Jiang Y, Yu H, Wang X. Qin Q, et al. Cell Mol Immunol. 2021 Apr;18(4):992-1004. doi: 10.1038/s41423-020-00525-3. Epub 2020 Sep 8. Cell Mol Immunol. 2021. PMID: 32901127 Free PMC article. - A comprehensive map of the toll-like receptor signaling network.
Oda K, Kitano H. Oda K, et al. Mol Syst Biol. 2006;2:2006.0015. doi: 10.1038/msb4100057. Epub 2006 Apr 18. Mol Syst Biol. 2006. PMID: 16738560 Free PMC article. Review. - Regulation of RKIP binding to the N-region of the Raf-1 kinase.
Park S, Rath O, Beach S, Xiang X, Kelly SM, Luo Z, Kolch W, Yeung KC. Park S, et al. FEBS Lett. 2006 Nov 27;580(27):6405-12. doi: 10.1016/j.febslet.2006.10.054. Epub 2006 Nov 3. FEBS Lett. 2006. PMID: 17097642 Free PMC article. - What signals are generated by anti-CD20 antibody therapy?
Bonavida B. Bonavida B. Curr Hematol Malig Rep. 2006 Dec;1(4):205-13. doi: 10.1007/s11899-006-0001-z. Curr Hematol Malig Rep. 2006. PMID: 20425315 Review.
References
- Baldwin A S., Jr The NF-κ B and I κ B proteins: new discoveries and insights. Annu Rev Immunol. 1996;14:649–683. - PubMed
- Banfield M J, Barker J J, Perry A C, Brady R L. Function from structure? The crystal structure of human phosphatidylethanolamine-binding protein suggests a role in membrane signal transduction. Structure. 1998;6:1245–1254. - PubMed
- Banfield M J, Brady R L. The structure of Antirrhinum centroradialis protein (CEN) suggests a role as a kinase regulator. J Mol Biol. 2000;297:1159–1170. - PubMed
- Bradley D, Carpenter R, Copsey L, Vincent C, Rothstein S, Coen E. Control of inflorescence architecture in Antirrhinum. Nature. 1996;379:791–797. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P20RR15578/RR/NCRR NIH HHS/United States
- P20 RR015578/RR/NCRR NIH HHS/United States
- R01 GM64767/GM/NIGMS NIH HHS/United States
- R01 GM064767/GM/NIGMS NIH HHS/United States
- R01 GM041690/GM/NIGMS NIH HHS/United States
- GM 41690/GM/NIGMS NIH HHS/United States
- R01 GM55435/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous